8JV7
 
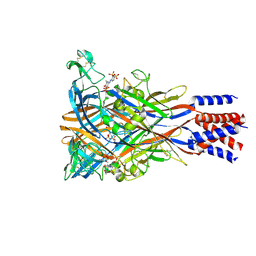 | | Cryo-EM structure of the panda P2X7 receptor in complex with PPADS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(E)-[4-methanoyl-6-methyl-5-oxidanyl-3-(phosphonooxymethyl)pyridin-2-yl]diazenyl]benzene-1,3-disulfonic acid, P2X purinoceptor | | Authors: | Sheng, D, Hattori, M. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the orthosteric inhibition of P2X receptors by non-ATP analog antagonists.
Elife, 12, 2024
|
|
1PUL
 
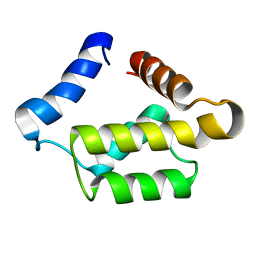 | | Solution structure for the 21KDa caenorhabditis elegans protein CE32E8.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR33 | | Descriptor: | Hypothetical protein C32E8.3 in chromosome I | | Authors: | Tejero, R, Aramini, J.M, Swapna, G.V.T, Monleon, D, Chiang, Y, Macapagal, D, Gunsalus, K.C, Kim, S, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-06-25 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone 1H, 15N and 13C assignments for the 21 kDa Caenorhabditis elegans homologue of "brain-specific" protein.
J.Biomol.Nmr, 28, 2004
|
|
8W1V
 
 | | The beta2 adrenergic receptor bound to a bitopic ligand | | Descriptor: | (2S)-1-[(3-{1-[4-(4-{(2S)-2-hydroxy-3-[(propan-2-yl)amino]propoxy}phenyl)butyl]-1H-1,2,3-triazol-4-yl}propyl)amino]-3-(2-propylphenoxy)propan-2-ol, Beta-2 adrenergic receptor,Endolysin, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Gaiser, B, Danielsen, M, Xu, X, Jorgensen, K, Fronik, P, Marcher-Rorsted, E, Wrobe, T, Hirata, K, Liu, X, Mathiesen, J, Pedersen, D. | | Deposit date: | 2024-02-19 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bitopic Ligands Support the Presence of a Metastable Binding Site at the beta 2 Adrenergic Receptor.
J.Med.Chem., 67, 2024
|
|
8JV8
 
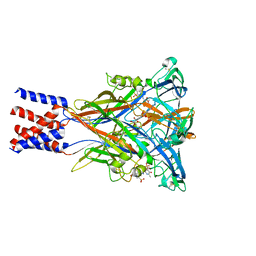 | | Cryo-EM structure of the panda P2X7 receptor in complex with PPNDS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, P2X purinoceptor | | Authors: | Sheng, D, Hattori, M. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural insights into the orthosteric inhibition of P2X receptors by non-ATP analog antagonists.
Elife, 12, 2024
|
|
1QMA
 
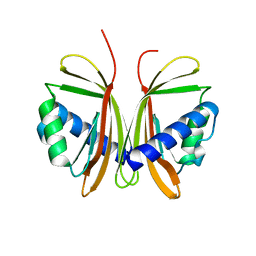 | | Nuclear Transport Factor 2 (NTF2) W7A mutant | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Bayliss, R, Ribbeck, K, Akin, D, Kent, H.M, Feldherr, C.M, Gorlich, D, Stewart, M.J. | | Deposit date: | 1999-09-23 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction Betweeen Ntf2 and Xfxfg-Containing Nucleoporins is Required to Mediate Nuclear Import of Ran-Gdp
J.Mol.Biol., 293, 1999
|
|
4WTD
 
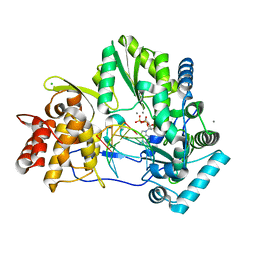 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS AND DELTA8 BETA HAIRPIN LOOP DELETION IN COMPLEX WITH ADP, MN2+ AND SYMMETRICAL PRIMER TEMPLATE 5'-AUAAAUUU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Edwards, T.E, Fox III, D, Appleby, T.C. | | Deposit date: | 2014-10-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
7QO4
 
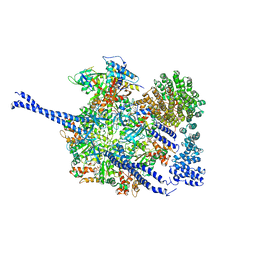 | | 26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn1, Ubp6, and UbVS) | | Descriptor: | 26S proteasome regulatory subunit RPN1, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
4FL1
 
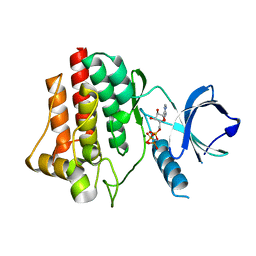 | | Structural and Biophysical Characterization of the Syk Activation Switch | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tyrosine-protein kinase SYK | | Authors: | Graedler, U, Schwarz, D, Dresing, V, Musil, M, Bomke, J, Frech, M, Jaekel, S, Rysiok, T, Mueller-Pompalla, D, Wegener, A. | | Deposit date: | 2012-06-14 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and biophysical characterization of the syk activation switch.
J.Mol.Biol., 425, 2013
|
|
7AUX
 
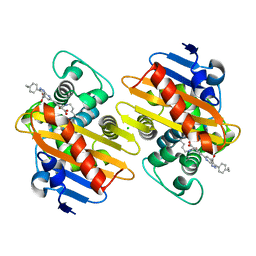 | | Crystal structure of OXA-48 beta-lactamase in the complex with the inhbitor ID2 | | Descriptor: | 6-(4-carboxyphenyl)-3-(4-ethylphenyl)-2~{H}-pyrazolo[3,4-b]pyridine-4-carboxylic acid, Beta-lactamase, CHLORIDE ION | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Garofalo, B, Ombrato, R. | | Deposit date: | 2020-11-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Novel Chemical Series of OXA-48 beta-Lactamase Inhibitors by High-Throughput Screening.
Pharmaceuticals, 14, 2021
|
|
7AW5
 
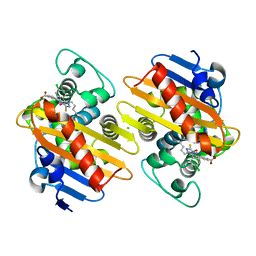 | | Crystal structure of OXA-48 beta-lactamase in the complex with the inhibitor ID3 | | Descriptor: | 4-[(~{E})-[3-(4-chlorophenyl)-5-sulfanylidene-1~{H}-1,2,4-triazol-4-yl]iminomethyl]benzoic acid, Beta-lactamase, CHLORIDE ION | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Garofalo, B, Ombrato, R. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Novel Chemical Series of OXA-48 beta-Lactamase Inhibitors by High-Throughput Screening.
Pharmaceuticals, 14, 2021
|
|
7AT5
 
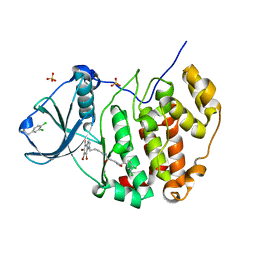 | | Structure of protein kinase ck2 catalytic subunit (csnk2a1 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, Casein kinase II subunit alpha, ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
7AT9
 
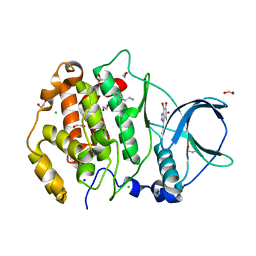 | | Structure of protein kinase ck2 catalytic subunit (csnk2a2 gene product) in complex with the ATP-competitive inhibitor MB002 and the alphaD-pocket ligand 3,4-dichlorophenethylamine | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol, ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
7ATV
 
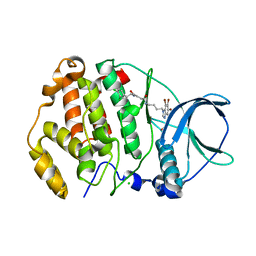 | | Structure of protein kinase ck2 catalytic subunit (csnk2a2 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha', ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-31 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
8OTO
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with AMP | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8OSX
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with ATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
1R7D
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 51 structures, sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
8OTR
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with SAM analog BDH 33959089 | | Descriptor: | (2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-N-(1-methylpiperidin-4-yl)-3,4-bis(oxidanyl)oxolane-2-carboxamide, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8OT0
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with MTA and glycine | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Falke, S. | | Deposit date: | 2023-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8WGN
 
 | | Human dihydrofolate reductase (HsDHFR) complexed with NADPH and LA1 | | Descriptor: | (3~{E})-3-[[2-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]oxypropoxy]phenyl]methylidene]oxolan-2-one, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vanichtanankul, J, Saeyang, T, Vitsupakorn, D, Decharuangsilp, S, Arwon, U, Yuthavong, Y, Kamchonwongpaisan, S. | | Deposit date: | 2023-09-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexible 2,4-diaminopyrimidine bearing a butyrolactone as Plasmodium falciparum dihydrofolate reductase inhibitors.
Bioorg.Chem., 153, 2024
|
|
8WGM
 
 | | Quadruple mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS V1/S, N51I+C59R+S108N+I164L) complexed with LA1, NADPH and dUMP | | Descriptor: | (3~{E})-3-[[2-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]oxypropoxy]phenyl]methylidene]oxolan-2-one, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Saeyang, T, Vitsupakorn, D, Arwon, U, Decharuangsilp, S, Yuthavong, Y, Kamchonwongpaisan, S. | | Deposit date: | 2023-09-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Flexible 2,4-diaminopyrimidine bearing a butyrolactone as Plasmodium falciparum dihydrofolate reductase inhibitors.
Bioorg.Chem., 153, 2024
|
|
6ZS5
 
 | | 3.5 A cryo-EM structure of human uromodulin filament core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
1SCZ
 
 | | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase | | Descriptor: | Dihydrolipoamide Succinyltransferase | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Tsao, J, Johnson, D, Huang, W.-Y, Pruett, P, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, Z, Lu, S, DeLucas, L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase
To be Published
|
|
7AWT
 
 | | E. coli NADH quinone oxidoreductase hydrophilic arm | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Schimpf, J, Grishkovskaya, I, Haselbach, D, Friedrich, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structure of the peripheral arm of a minimalistic respiratory complex I.
Structure, 30, 2022
|
|
7AIL
 
 | |
7AGJ
 
 | | Ribonucleotide Reductase R1 protein from Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Rehling, D, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2020-09-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Investigation of Class I Ribonucleotide Reductase from the Hyperthermophile Aquifex aeolicus.
Biochemistry, 61, 2022
|
|
