7NR8
 
 | | Discovery of ASTX029, a clinical candidate which modulates the phosphorylation and catalytic activity of ERK1/2 | | Descriptor: | (2~{R})-2-[5-[5-chloranyl-2-(oxan-4-ylamino)pyrimidin-4-yl]-3-oxidanylidene-1~{H}-isoindol-2-yl]-~{N}-[(1~{S})-1-[6-(4-methylpiperazin-1-yl)pyridin-2-yl]-2-oxidanyl-ethyl]propanamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | O'Reilly, M, Cleasby, A. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.627 Å) | | Cite: | Discovery of ASTX029, A Clinical Candidate Which Modulates the Phosphorylation and Catalytic Activity of ERK1/2.
J.Med.Chem., 64, 2021
|
|
7NR5
 
 | | Discovery of ASTX029, a clinical candidate which modulates the phosphorylation and catalytic activity of ERK1/2 | | Descriptor: | (2~{R})-2-[5-[5-chloranyl-2-[(2-methyl-1,2,3-triazol-4-yl)amino]pyrimidin-4-yl]-3-oxidanylidene-1~{H}-isoindol-2-yl]-~{N}-[(1~{S})-1-(3-fluoranyl-5-methoxy-phenyl)-2-oxidanyl-ethyl]propanamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Discovery of ASTX029, A Clinical Candidate Which Modulates the Phosphorylation and Catalytic Activity of ERK1/2.
J.Med.Chem., 64, 2021
|
|
5LY9
 
 | | Structure of MITat 1.1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Variant surface glycoprotein MITAT 1.1, ... | | Authors: | Schaefer, C, Bartossek, T, Jones, N, Kuper, J, Kisker, C, Engstler, M. | | Deposit date: | 2016-09-26 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the shielding function of the dynamic trypanosome variant surface glycoprotein coat.
Nat Microbiol, 2, 2017
|
|
6MT1
 
 | |
6JPP
 
 | |
6MT2
 
 | |
7BGU
 
 | | Mason-Pfizer Monkey Virus Protease mutant C7A/D26N/C106A in complex with peptidomimetic inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PENTAETHYLENE GLYCOL, peptidomimetic inhibitor | | Authors: | Wosicki, S, Gilski, M, Kazmierczyk, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2021-01-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Crystal structures of inhibitor complexes of M-PMV protease with visible flap loops.
Protein Sci., 30, 2021
|
|
7ONT
 
 | | PARP1 catalytic domain in complex with a selective pyridine carboxamide-based inhibitor (compound 22) | | Descriptor: | 5-[4-[(3-ethyl-2-oxidanylidene-1~{H}-quinolin-7-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M.J, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
6JLO
 
 | | XFEL structure of cyanobacterial photosystem II (2F state, dataset2) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
6JLL
 
 | | XFEL structure of cyanobacterial photosystem II (2F state, dataset1) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An oxyl/oxo mechanism for oxygen-oxygen coupling in PSII revealed by an x-ray free-electron laser.
Science, 366, 2019
|
|
7QQE
 
 | | Nuclear factor one X - NFIX in P41212 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nuclear factor 1 X-type, ZINC ION | | Authors: | Lapi, M, Chaves-Sanjuan, A, Gourlay, L.J, Tiberi, M, Polentarutti, M, Demitri, N, Bais, G, Nardini, M. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nuclear factor one X - NFIX in P41212
To Be Published
|
|
7QQD
 
 | | Nuclear factor one X - NFIX in P21 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NFI binding site (forward), NFI binding site (reverse), ... | | Authors: | Lapi, M, Chaves-Sanjuan, A, Gourlay, L.J, Tiberi, M, Polentarutti, M, Demitri, N, Bais, G, Nardini, M. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nuclear factor one X - NFIX in P41212
To Be Published
|
|
7QRM
 
 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.7 A resolution | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Sarewicz, M, Szwalec, M, Indyka, P, Rawski, M, Pintscher, S, Pietras, R, Mielecki, B, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-01-11 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
8JZS
 
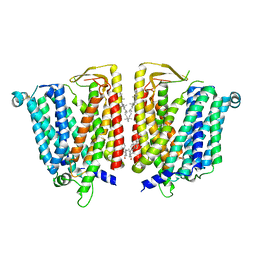 | | Outward-facing SLC15A4 dimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, lysosomal transporter | | Authors: | Zhang, S.S, Chen, X.D, Xie, M. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for recruitment of TASL by SLC15A4 in human endolysosomal TLR signaling.
Nat Commun, 14, 2023
|
|
8JZU
 
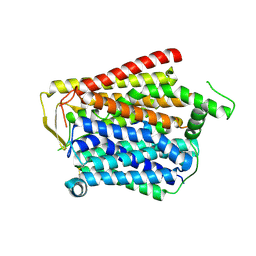 | | SLC15A4_TASL complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, TLR adapter,Green fluorescent protein, TSLAA-EGPF tag fusion protein | | Authors: | Zhang, S.S, Chen, X.D, Xie, M. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for recruitment of TASL by SLC15A4 in human endolysosomal TLR signaling.
Nat Commun, 14, 2023
|
|
7OLY
 
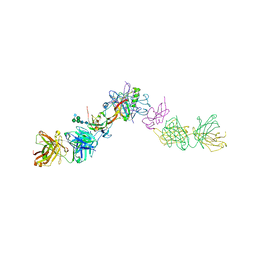 | | Structure of activin A in complex with an ActRIIB-Alk4 fusion reveal insight into activin receptor interactions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-1B, ... | | Authors: | Hakansson, M, Rose, N.C, Castonguay, R, Logan, D.T, Krishnan, L. | | Deposit date: | 2021-05-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.265 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
8JZR
 
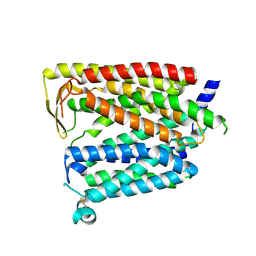 | | Outward_facing SLC15A4 monomer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, lysosomal transporter,ALFA tag | | Authors: | Zhang, S.S, Chen, X.D, Xie, M. | | Deposit date: | 2023-07-06 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for recruitment of TASL by SLC15A4 in human endolysosomal TLR signaling.
Nat Commun, 14, 2023
|
|
6TRJ
 
 | | LEDGF/p75 IBD dimer | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Kugler, M, Brynda, J. | | Deposit date: | 2019-12-19 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fine-tuning of the LEDGF/p75 interaction network by dimerization
Structure
|
|
8B75
 
 | | CRYSTAL STRUCTURE OF HUMAN SOLUBLE ADENYLYL CYCLASE IN COMPLEX WITH THE INHIBITOR TDI-011861 | | Descriptor: | Adenylate cyclase type 10, [(3~{R})-4-[2-[2-[[3-(2-azanyl-6-chloranyl-pyrimidin-4-yl)-1-[bis(fluoranyl)methyl]pyrazol-4-yl]methyl]phenoxy]ethyl]morpholin-3-yl]methanol | | Authors: | Steegborn, C, Kehr, M. | | Deposit date: | 2022-09-28 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design, Synthesis, and Pharmacological Evaluation of Second-Generation Soluble Adenylyl Cyclase (sAC, ADCY10) Inhibitors with Slow Dissociation Rates.
J.Med.Chem., 65, 2022
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
8SUV
 
 | | CHIP-TPR in complex with the C-terminus of CHIC2 | | Descriptor: | Cysteine-rich hydrophobic domain-containing protein 2, E3 ubiquitin-protein ligase CHIP, SULFATE ION | | Authors: | Cupo, A.R, McDermott, L.E, DeSilva, A.R, Callahan, M, Nix, J.C, Gestwicki, J.E, Page, R.C. | | Deposit date: | 2023-05-13 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Interaction with the membrane-anchored protein CHIC2 constrains the ubiquitin ligase activity of CHIP
Biorxiv, 2023
|
|
7OTC
 
 | | Cryo-EM structure of an Escherichia coli 70S ribosome in complex with elongation factor G and the antibiotic Argyrin B | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Wieland, M, Koller, T.O, Wilson, D.N. | | Deposit date: | 2021-06-10 | | Release date: | 2022-05-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The cyclic octapeptide antibiotic argyrin B inhibits translation by trapping EF-G on the ribosome during translocation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7P9O
 
 | | Structure of E.coli RlmJ in complex with a SAM analogue (CA) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]amino}-5'-deoxyadenosine, Ribosomal RNA large subunit methyltransferase J | | Authors: | Meynier, V, Catala, M, Oerum, S, Barraud, P, Tisne, C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Synthesis of RNA-cofactor conjugates and structural exploration of RNA recognition by an m6A RNA methyltransferase.
Nucleic Acids Res., 50, 2022
|
|
7OMO
 
 | |
7P8Q
 
 | | Structure of E.coli RlmJ in complex with an RNA conjugate (GA-SAM) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](3-aminopropyl)amino}-5'-deoxyadenosine, RNA conjugate (GA-SAM), Ribosomal RNA large subunit methyltransferase J | | Authors: | Meynier, V, Catala, M, Oerum, S, Barraud, P, Tisne, C. | | Deposit date: | 2021-07-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Synthesis of RNA-cofactor conjugates and structural exploration of RNA recognition by an m6A RNA methyltransferase.
Nucleic Acids Res., 50, 2022
|
|
