6PBN
 
 | | Pseudopaline Dehydrogenase with (R)-Pseudopaline Soaked 1 hour | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, N-[(3S)-3-amino-3-carboxypropyl]-L-histidine, ... | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
6PBM
 
 | | Pseudopaline Dehydrogenase with NADP+ bound | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pseudopaline Dehydrogenase | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
6PBT
 
 | | Pseudopaline Dehydrogenase with (R)-Pseudopaline Soaked 2 hours | | Descriptor: | 1,2-ETHANEDIOL, N-[(3S)-3-amino-3-carboxypropyl]-L-histidine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
8DEB
 
 | | Bacteroides fragilis carboxyspermidine dehydrogenase | | Descriptor: | Carboxyspermidine dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | McFarlane, J.S, Bouchey, S, Dodd, J. | | Deposit date: | 2022-06-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Kinetic and structural characterization of carboxyspermidine dehydrogenase of polyamine biosynthesis.
J.Biol.Chem., 299, 2023
|
|
6PBP
 
 | | Pseudopaline Dehydrogenase with (S)-Pseudopaline Soaked 1 hour | | Descriptor: | 1,2-ETHANEDIOL, N-[(1S)-1-carboxy-3-{[(1S)-1-carboxy-2-(1H-imidazol-5-yl)ethyl]amino}propyl]-L-glutamic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
6NN8
 
 | | The structure of human liver pyruvate kinase, hLPYK-S531E | | Descriptor: | 1,2-ETHANEDIOL, Pyruvate kinase PKLR | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NN4
 
 | | The structure of human liver pyruvate kinase, hLPYK-D499N, in complex with Fru-1,6-BP | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, PHOSPHOENOLPYRUVATE, ... | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NN5
 
 | | The structure of human liver pyruvate kinase, hLPYK-W527H | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pyruvate kinase PKLR | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NN7
 
 | | The structure of human liver pyruvate kinase, hLPYK-GGG | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, GLYCEROL, ... | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7UEH
 
 | | Pyruvate kinase from Zymomonas mobilis | | Descriptor: | GLYCEROL, PHOSPHATE ION, Pyruvate kinase | | Authors: | McFarlane, J.S, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.4 angstrom structure of Zymomonas mobilis pyruvate kinase: Implications for stability and regulation.
Arch.Biochem.Biophys., 744, 2023
|
|
6C4N
 
 | | Pseudopaline dehydrogenase (PaODH) - NADP+ bound | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pseudopaline dehydrogenase | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6C4L
 
 | | Yersinopine dehydrogenase (YpODH) - Apo | | Descriptor: | Yersinopine dehydrogenase | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6C4M
 
 | | Yersinopine dehydrogenase (YpODH) - NADP+ bound | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Yersinopine dehydrogenase | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6C4R
 
 | | Staphylopine dehydrogenase (SaODH) - Apo | | Descriptor: | GLYCEROL, SULFATE ION, Staphylopine dehydrogenase | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6C4T
 
 | | Staphylopine dehydrogenase (SaODH) - NADP+ bound | | Descriptor: | GLYCEROL, Staphylopine dehydrogenase, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6DQO
 
 | |
6DQI
 
 | |
6DQP
 
 | |
8CDQ
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to the inhibitor KNX-002 and Mg.ATP-gamma-S | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-methoxyphenyl)-~{N}-[(3-thiophen-2-yl-1~{H}-pyrazol-4-yl)methyl]cyclopropan-1-amine, GLYCEROL, ... | | Authors: | Moussaoui, D, Robblee, J.P, Robert-Paganin, J, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Schaletzky, J, Wehri, E, Mueller-Dieckmann, C, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2023-01-31 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
1GWY
 
 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II | | Descriptor: | STICHOLYSIN II, SULFATE ION | | Authors: | Mancheno, J.M, Martin-Benito, J, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-03-26 | | Release date: | 2003-06-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
7ONS
 
 | | PARP1 catalytic domain in complex with isoquinolone-based inhibitor (compound 16) | | Descriptor: | 7-[[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]methyl]-3-ethyl-1~{H}-quinolin-2-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
7ONT
 
 | | PARP1 catalytic domain in complex with a selective pyridine carboxamide-based inhibitor (compound 22) | | Descriptor: | 5-[4-[(3-ethyl-2-oxidanylidene-1~{H}-quinolin-7-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M.J, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
7ONR
 
 | | PARP1 catalytic domain in complex with 8-chloroquinazolinone-based inhibitor (compound 9) | | Descriptor: | 8-chloranyl-2-[3-[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]propyl]-3~{H}-quinazolin-4-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
3EML
 
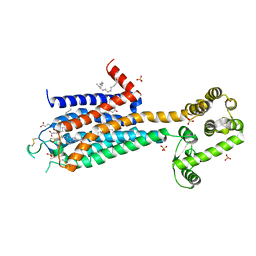 | | The 2.6 A Crystal Structure of a Human A2A Adenosine Receptor bound to ZM241385. | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Human Adenosine A2A receptor/T4 lysozyme chimera, STEARIC ACID, ... | | Authors: | Jaakola, V.-P, Griffith, M.T, Hanson, M.A, Cherezov, V, Chien, E.Y.T, Lane, J.R, Ijzerman, A.P, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2008-09-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist.
Science, 322, 2008
|
|
1AFP
 
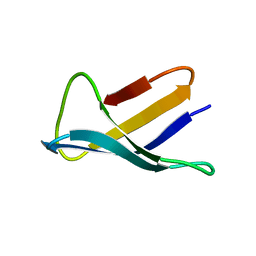 | | SOLUTION STRUCTURE OF THE ANTIFUNGAL PROTEIN FROM ASPERGILLUS GIGANTEUS. EVIDENCE FOR DISULPHIDE CONFIGURATIONAL ISOMERISM | | Descriptor: | ANTIFUNGAL PROTEIN FROM ASPERGILLUS GIGANTEUS | | Authors: | Campos-Olivas, R, Bruix, M, Santoro, J, Lacadena, J, Del Pozo, A.M, Gavilanes, J.G, Rico, M. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the antifungal protein from Aspergillus giganteus: evidence for cysteine pairing isomerism.
Biochemistry, 34, 1995
|
|
