8Y94
 
 | |
8Y95
 
 | | Structure of NET-NE in Occluded state | | Descriptor: | CHLORIDE ION, Noradrenaline, SODIUM ION, ... | | Authors: | Zhang, H, Xu, H.E, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8ZJE
 
 | | Cryo-EM structure of kisspeptin receptor bound to TAK-448 | | Descriptor: | ACE-DTY-HYP-ASN-THR-PHE-XZA-LEU-NMM-TRP-NH2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shen, S, Liu, H, Xu, H.E. | | Deposit date: | 2024-05-14 | | Release date: | 2024-07-03 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for hormone recognition and distinctive Gq protein coupling by the kisspeptin receptor.
Cell Rep, 43, 2024
|
|
8J6I
 
 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound MK-6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-04-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
8J6L
 
 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound niacin | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-04-03 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
8JR9
 
 | | Small molecule agonist (PCO371) bound to human parathyroid hormone receptor type 1 (PTH1R) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, He, X, Shan, H, Li, J, Wang, K, Li, Y, Hu, W, Wu, K, Shen, J, Xu, H.E. | | Deposit date: | 2023-06-16 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Conserved class B GPCR activation by a biased intracellular agonist.
Nature, 621, 2023
|
|
8J6J
 
 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound with GSK256073 | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
8YW5
 
 | | Cryo-EM structure of the retatrutide-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, W.Z, Zhou, Q.T, Cong, Z.T, Yuan, Q.N, Li, W.X, Zhao, F.H, Xu, H.E, Zhao, L.H, Yang, D.H, Wang, M.W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-08-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into the triple agonism at GLP-1R, GIPR and GCGR manifested by retatrutide.
Cell Discov, 10, 2024
|
|
8YW3
 
 | | Cryo-EM structure of the retatrutide-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, W.Z, Zhou, Q.T, Cong, Z.T, Yuan, Q.N, Li, W.X, Zhao, F.H, Xu, H.E, Zhao, L.H, Yang, D.H, Wang, M.W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural insights into the triple agonism at GLP-1R, GIPR and GCGR manifested by retatrutide.
Cell Discov, 10, 2024
|
|
7F16
 
 | | Cryo-EM structure of parathyroid hormone receptor type 2 in complex with a tuberoinfundibular peptide of 39 residues and G protein | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, X, Cheng, X, Zhao, L.H, Wang, Y, Ye, C, Zou, X, Dai, A, Cong, Z, Chen, J, Zhou, Q, Xia, T, Jiang, H, Xu, H.E, Yang, D, Wang, M.W. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular insights into differentiated ligand recognition of the human parathyroid hormone receptor 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EZH
 
 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gi complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
7EZK
 
 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gs complex | | Descriptor: | Chimera of Guanine nucleotide-binding protein G(i) subunit alpha-1 and Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
7EZM
 
 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gq complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
7F4H
 
 | | Cryo-EM structure of afamelanotide-bound melanocortin-1 receptor in complex with Gs protein, Nb35 and scFv16 | | Descriptor: | Afamelanotide, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7F4I
 
 | | Cryo-EM structure of SHU9119-bound melanocortin-1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7F4D
 
 | | Cryo-EM structure of alpha-MSH-bound melanocortin-1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7F4F
 
 | | Cryo-EM structure of afamelanotide-bound melanocortin-1 receptor in complex with Gs protein and scFv16 | | Descriptor: | Afamelanotide, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7EPT
 
 | | Structural basis for the tethered peptide activation of adhesion GPCRs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ping, Y.-Q, Xiao, P, Yang, F, Zhao, R.-J, Guo, S.-C, Yan, X, Wu, X, Liebscher, I, Xu, H.E, Sun, J.-P. | | Deposit date: | 2021-04-27 | | Release date: | 2022-05-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
8YW4
 
 | | Cryo-EM structure of the retatrutide-bound human GIPR-Gs complex | | Descriptor: | Gastric inhibitory polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, W.Z, Zhou, Q.T, Cong, Z.T, Yuan, Q.N, Li, W.X, Zhao, F.H, Xu, H.E, Zhao, L.H, Yang, D.H, Wang, M.W. | | Deposit date: | 2024-03-29 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into the triple agonism at GLP-1R, GIPR and GCGR manifested by retatrutide.
Cell Discov, 10, 2024
|
|
7D68
 
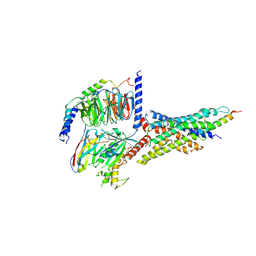 | | Cryo-EM structure of the human glucagon-like peptide-2 receptor-Gs protein complex | | Descriptor: | Glucagon-like peptide 2 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, W, Chen, L, Zhou, Q, Zhao, L, Zhang, H, Cong, Z, Shen, D, Zhao, F, Zhou, F, Cai, X, Chen, Y, Zhou, Y, Gadgaard, S, van der Velden, W.J, Zhao, S, Jiang, Y, Rosenkilde, M.M, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor.
Cell Res., 30, 2020
|
|
8ZPT
 
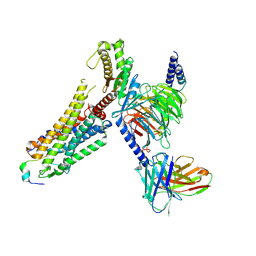 | | Cryo-EM structure of prolactin-releasing peptide recognition with Gq | | Descriptor: | Guanine nucleotide-binding protein G(324) subunit alpha-1,, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, L, Li, Y, Yuan, Q, Xu, H.E. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-25 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Molecular mechanism of prolactin-releasing peptide recognition and signaling via its G protein-coupled receptor.
Cell Discov, 10, 2024
|
|
8ZPS
 
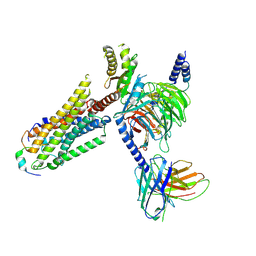 | | Cryo-EM structure of prolactin-releasing peptide recognition with Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha-1, ... | | Authors: | Zhao, L, Li, Y, Yuan, Q, Xu, H.E. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Molecular mechanism of prolactin-releasing peptide recognition and signaling via its G protein-coupled receptor.
Cell Discov, 10, 2024
|
|
8ZYU
 
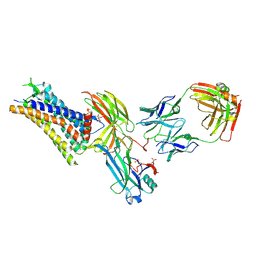 | | Cryo-EM structure of neurotensin receptor 1 in complex with beta-arrestin1 and SBI-553 (complex 1) | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Beta-arrestin-1, Fab30 heavy chain, ... | | Authors: | Sun, D, Li, X, Yuan, Q, Yin, W, Xu, H.E, Tian, C. | | Deposit date: | 2024-06-18 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Molecular mechanism of the arrestin-biased agonism of neurotensin receptor 1 by an intracellular allosteric modulator.
Cell Res., 35, 2025
|
|
8ZYT
 
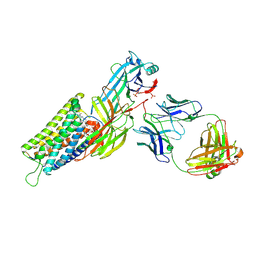 | | Cryo-EM structure of neurotensin receptor 1 in complex with beta-arrestin1 and SBI-553 (complex 3) | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Beta-arrestin-1, Fab30 heavy chain, ... | | Authors: | Sun, D, Li, X, Yuan, Q, Yin, W, Xu, H.E, Tian, C. | | Deposit date: | 2024-06-18 | | Release date: | 2025-04-09 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Molecular mechanism of the arrestin-biased agonism of neurotensin receptor 1 by an intracellular allosteric modulator.
Cell Res., 35, 2025
|
|
8ZYY
 
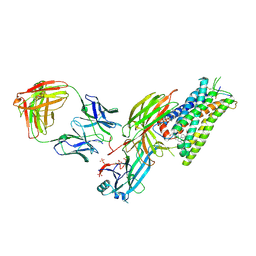 | | Cryo-EM structure of neurotensin receptor 1 in complex with beta-arrestin1 and SBI-553 (complex 2) | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Beta-arrestin-1, Fab30 heavy chain, ... | | Authors: | Sun, D, Li, X, Yuan, Q, Yin, W, Xu, H.E, Tian, C. | | Deposit date: | 2024-06-18 | | Release date: | 2025-04-09 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Molecular mechanism of the arrestin-biased agonism of neurotensin receptor 1 by an intracellular allosteric modulator.
Cell Res., 35, 2025
|
|
