8R8R
 
 | | Cryo-EM structure of the human mPSF with PAPOA C-terminus peptide (PAPOAc) | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*AP*AP*UP*AP*AP*A)-3'), ... | | Authors: | Todesca, S, Sandmeir, F, Keidel, A, Conti, E. | | Deposit date: | 2023-11-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Molecular basis of human poly(A) polymerase recruitment by mPSF.
Rna, 30, 2024
|
|
8PUH
 
 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-CTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 2024
|
|
8PUG
 
 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 2024
|
|
8OST
 
 | | Structure of human terminal uridylyltransferase 4 (TUT4, ZCCHC11) in complex with pre-let7g miRNA and Lin28A | | Descriptor: | Protein lin-28 homolog A, Terminal uridylyltransferase 4, ZINC ION, ... | | Authors: | Gilbert, R.J, Yi, G, Ye, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 2024
|
|
8OPT
 
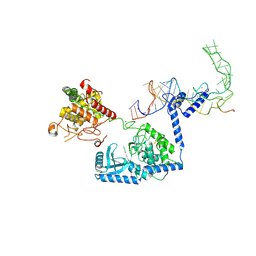 | |
8OPS
 
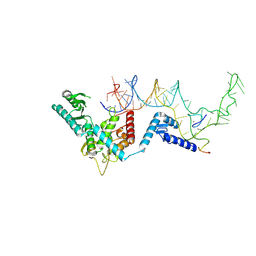 | |
8OPP
 
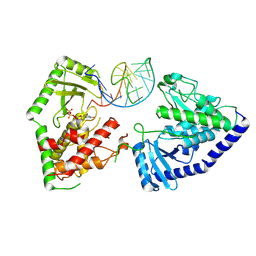 | | Structure of human terminal uridylyltransferase 7 (hTUT7/ZCCHC6) bound with pre-let7g miRNA and UTPalphaS | | Descriptor: | RNA (25-MER), Terminal uridylyltransferase 7, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Yi, G, Ye, M, Gilbert, R.J. | | Deposit date: | 2023-04-07 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 2024
|
|
8OEF
 
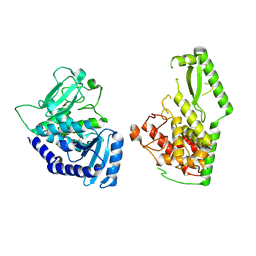 | |
8E3Q
 
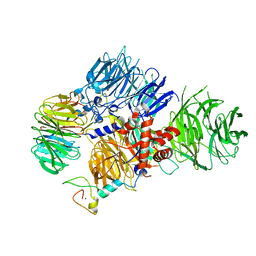 | | CRYO-EM STRUCTURE OF the human MPSF | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, ZINC ION, ... | | Authors: | Gutierrez, P.A, Wei, J, Sun, Y, Tong, L. | | Deposit date: | 2022-08-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Molecular basis for the recognition of the AUUAAA polyadenylation signal by mPSF.
Rna, 28, 2022
|
|
8E3I
 
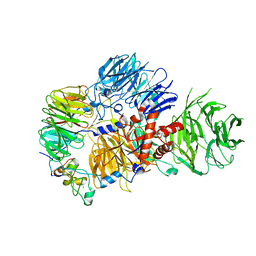 | | CRYO-EM STRUCTURE OF the human MPSF IN COMPLEX WITH THE AUUAAA poly(A) signal | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*CP*AP*UP*UP*AP*AP*AP*CP*AP*AP*C)-3'), ... | | Authors: | Gutierrez, P.A, Wei, J, Sun, Y, Tong, L. | | Deposit date: | 2022-08-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Molecular basis for the recognition of the AUUAAA polyadenylation signal by mPSF.
Rna, 28, 2022
|
|
8C9M
 
 | | HERV-K Gag immature lattice | | Descriptor: | Gag protein | | Authors: | Krebs, A.-S, Liu, H.-F, Zhou, Y, Rey, J.S, Levintov, L, Perilla, J.R, Bartesaghi, A, Zhang, P. | | Deposit date: | 2023-01-23 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and conservation of an immature human endogenous retrovirus.
Biorxiv, 2023
|
|
7Z52
 
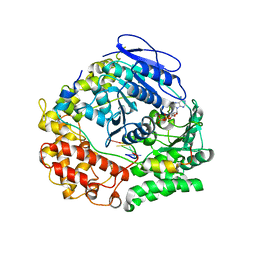 | | Human NEXT dimer - focused reconstruction of the single MTR4 | | Descriptor: | Exosome RNA helicase MTR4, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(P*UP*UP*UP*UP*U)-3'), ... | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
7Z4Z
 
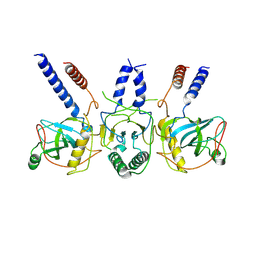 | | Human NEXT dimer - focused reconstruction of the dimerization module | | Descriptor: | Exosome RNA helicase MTR4, Zinc finger CCHC domain-containing protein 8 | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
7Z4Y
 
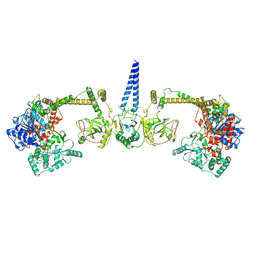 | | Human NEXT dimer - overall reconstruction of the core complex | | Descriptor: | Exosome RNA helicase MTR4, Zinc finger CCHC domain-containing protein 8 | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
7W5Z
 
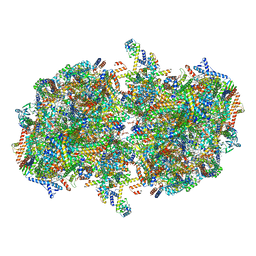 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial complex IV, composite dimer model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-oxoglutarate/malate carrier protein, CARDIOLIPIN, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Letts, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-04-06 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
7VPX
 
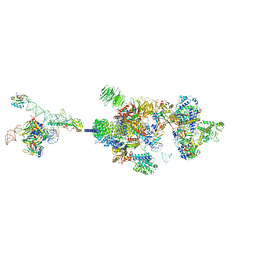 | | The cryo-EM structure of the human pre-A complex | | Descriptor: | 5SS, DnaJ homolog subfamily C member 8, PHD finger-like domain-containing protein 5A, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-10-18 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome.
Nat.Struct.Mol.Biol., 2024
|
|
7S7C
 
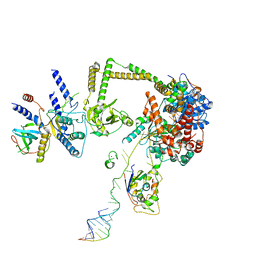 | |
7S7B
 
 | |
7C4C
 
 | |
7C4B
 
 | | The crystal structure of Trypanosoma brucei RNase D : UMP complex | | Descriptor: | CCHC-type domain-containing protein, MANGANESE (II) ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Gao, Y.Q, Gan, J.H. | | Deposit date: | 2020-05-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Nucleic Acids Res., 49, 2021
|
|
7C47
 
 | | The crystal structure of Trypanosoma brucei RNase D : CMP complex | | Descriptor: | CCHC-type domain-containing protein, CYTIDINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Gao, Y.Q, Gan, J.H. | | Deposit date: | 2020-05-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Nucleic Acids Res., 49, 2021
|
|
7C45
 
 | |
7C43
 
 | |
7C42
 
 | |
6URO
 
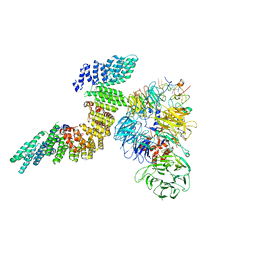 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-PAS RNA-CstF77 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, Cleavage stimulation factor subunit 3, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
