7C01
 
 | | Molecular basis for a potent human neutralizing antibody targeting SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CB6 heavy chain, CB6 light chain, ... | | Authors: | Shi, R, Qi, J, Wang, Q, Gao, F.G, Yan, J. | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2.
Nature, 584, 2020
|
|
7C2I
 
 | | Crystal structure of nsp16-nsp10 heterodimer from SARS-CoV-2 in complex with SAM (with additional SAM during crystallization) | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Lin, S, Chen, H, Ye, F, Chen, Z.M, Yang, F.L, Zheng, Y, Cao, Y, Qiao, J.X, Yang, S.Y, Lu, G.W. | | Deposit date: | 2020-05-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 2'-O-methylase and its implication on antiviral drug design.
Signal Transduct Target Ther, 5, 2020
|
|
6M1V
 
 | |
6YZ5
 
 | | H11-D4 complex with SARS-CoV-2 RBD | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Naismith, J.H, Huo, J, Mikolajek, H, Ward, P, Dumoux, M, Owens, R.J, Le Bas, A. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | H11-D4 complex with SARS-CoV-2 RBD
To Be Published
|
|
6YZ7
 
 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Cr3022, Antibody light chain, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published, 2020
|
|
6Z2M
 
 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 antibody, Spike glycoprotein, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-17 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published
|
|
6Z43
 
 | | Cryo-EM Structure of SARS-CoV-2 Spike : H11-D4 Nanobody Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Ruza, R.R, Duyvesteyn, H.M.E, Shah, P, Carrique, L, Ren, J, Malinauskas, T, Zhou, D, Stuart, D.I, Naismith, J.H. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for a potent neutralising single-domain antibody that blocks SARS-CoV-2 binding to its receptor ACE2
To Be Published
|
|
7BWJ
 
 | | crystal structure of SARS-CoV-2 antibody with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Wang, X, Ge, J. | | Deposit date: | 2020-04-14 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Human neutralizing antibodies elicited by SARS-CoV-2 infection.
Nature, 584, 2020
|
|
7BZF
 
 | | COVID-19 RNA-dependent RNA polymerase post-translocated catalytic complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (31-MER), ... | | Authors: | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | Deposit date: | 2020-04-27 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
7C2K
 
 | | COVID-19 RNA-dependent RNA polymerase pre-translocated catalytic complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (29-MER), ... | | Authors: | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | Deposit date: | 2020-05-07 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
6X6P
 
 | | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Herrera, N.G, Morano, N.C, Celikgil, A, Georgiev, G.I, Malonis, R, Lee, J.H, Tong, K, Vergnolle, O, Massimi, A, Yen, L.Y, Noble, A.J, Kopylov, M, Bonanno, J.B, Garrett-Thompson, S.C, Hayes, D.B, Brenowitz, M, Garforth, S.J, Eng, E.T, Lai, J.R, Almo, S.C. | | Deposit date: | 2020-05-28 | | Release date: | 2020-06-10 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis.
Biorxiv, 2020
|
|
7BYR
 
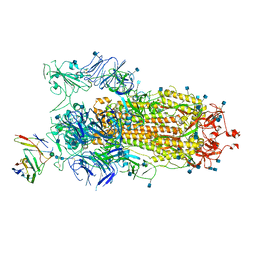 | | BD23-Fab in complex with the S ectodomain trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab23-Fab-Heavy Chain, ... | | Authors: | Zhu, Q, Wang, G, Xiao, J. | | Deposit date: | 2020-04-24 | | Release date: | 2020-06-10 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients' B Cells.
Cell, 182, 2020
|
|
6M5I
 
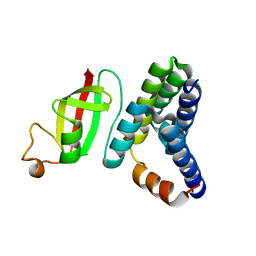 | | Crystal structure of 2019-nCoV nsp7-nsp8c complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Yan, L.M, Ge, J, Zhao, Y, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2020-03-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Crystal structure of 2019-nCoV nsp7-nsp8c complex
To Be Published
|
|
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | Descriptor: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | Descriptor: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
6XAA
 
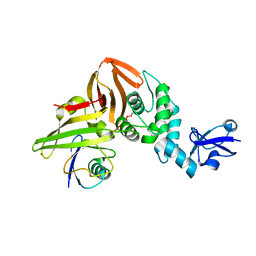 | | SARS CoV-2 PLpro in complex with ubiquitin propargylamide | | Descriptor: | Non-structural protein 3, Ubiquitin-propargylamide, ZINC ION | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
6XBG
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW246 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBH
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW247 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XBI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW248 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6XDC
 
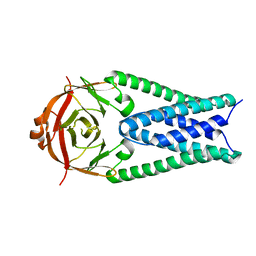 | |
6XDH
 
 | |
6Z4U
 
 | |
6XDG
 
 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies | | Descriptor: | REGN10933 antibody Fab fragment heavy chain, REGN10933 antibody Fab fragment light chain, REGN10987 antibody Fab fragment heavy chain, ... | | Authors: | Franklin, M.C, Saotome, K, Romero Hernandez, A, Zhou, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-06-24 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail.
Science, 369, 2020
|
|
6ZCZ
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in ternary complex with EY6A Fab and a nanobody. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EY6A heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZER
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|







