-Search query
-Search result
Showing 1 - 50 of 78 items for (author: zhu & ly)

EMDB-41363: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5
Method: single particle / : Yue H, Hunkeler M, Roy Burman SS, Fischer ES

PDB-8tl6: 
Cryo-EM structure of DDB1deltaB-DDA1-DCAF5
Method: single particle / : Yue H, Hunkeler M, Roy Burman SS, Fischer ES

EMDB-28198: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with LLNL-199
Method: single particle / : Binshtein E, Crowe JE

EMDB-28199: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with 2130-1-0114-112
Method: single particle / : Binshtein E, Crowe JE

PDB-8ekd: 
Cryo-EM map of SARS-CoV-2 Omicron BA.2 spike in complex with 2130-1-0114-112
Method: single particle / : Binshtein E, Crowe JE

EMDB-28156: 
Cryo-EM structure of human DNMT3B homo-tetramer (form I)
Method: single particle / : Lu JW, Song JK

EMDB-28157: 
Cryo-EM structure of human DNMT3B homo-tetramer (form II)
Method: single particle / : Lu JW, Song JK

EMDB-28158: 
Cryo-EM structure of human DNMT3B homo-trimer
Method: single particle / : Lu JW, Song JK

EMDB-28159: 
Cryo-EM structure of human DNMT3B homo-hexamer
Method: single particle / : Lu JW, Song JK

PDB-8eih: 
Cryo-EM structure of human DNMT3B homo-tetramer (form I)
Method: single particle / : Lu JW, Song JK

PDB-8eii: 
Cryo-EM structure of human DNMT3B homo-tetramer (form II)
Method: single particle / : Lu JW, Song JK

EMDB-33650: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7
Method: single particle / : Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AYY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Greaney AJ, Chen M, Au GG, Paradkar P, Peiris M, Chung AW, Bloom JD, Lye D, Lok SM, Wang LF

EMDB-33651: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface)
Method: single particle / : Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AYY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Greaney AJ, Chen M, Au GG, Paradkar P, Peiris M, Chung AW, Bloom JD, Lye D, Lok SM, Wang LF

PDB-7y71: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7
Method: single particle / : Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AYY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Greaney AJ, Chen M, Au GG, Paradkar P, Peiris M, Chung AW, Bloom JD, Lye D, Lok SM, Wang LF

PDB-7y72: 
SARS-CoV-2 spike glycoprotein trimer complexed with Fab fragment of anti-RBD antibody E7 (focused refinement on Fab-RBD interface)
Method: single particle / : Chia WN, Tan CW, Tan AWK, Young B, Starr TN, Lopez E, Fibriansah G, Barr J, Cheng S, Yeoh AYY, Yap WC, Lim BL, Ng TS, Sia WR, Zhu F, Chen S, Zhang J, Greaney AJ, Chen M, Au GG, Paradkar P, Peiris M, Chung AW, Bloom JD, Lye D, Lok SM, Wang LF

EMDB-40983: 
Negative stain EM assembly of MYC, JAZ, and NINJA complex
Method: single particle / : Zhou XE, Zhang Y, Zhou Y, He Q, Cao X, Kariapper L, Suino-Powell K, Zhu Y, Zhang F, Karsten M

PDB-8t2i: 
Negative stain EM assembly of MYC, JAZ, and NINJA complex
Method: single particle / : Zhou XE, Zhang Y, Zhou Y, He Q, Cao X, Kariapper L, Suino-Powell K, Zhu Y, Zhang F, Karsten M

EMDB-33448: 
Cryo-EM structure of Listeria monocytogenes man-PTS complexed with pediocin PA-1
Method: single particle / : Zeng JW, Wang JW, Zhu LY

EMDB-32252: 
Cryo-EM structure of human cohesin-CTCF-DNA complex
Method: single particle / : Shi ZB, Bai XC

PDB-7w1m: 
Cryo-EM structure of human cohesin-CTCF-DNA complex
Method: single particle / : Shi ZB, Bai XC, Yu H

EMDB-29735: 
Structure of nucleosome-bound Sirtuin 6 deacetylase
Method: single particle / : Chio US, Rechiche O, Bryll AR, Zhu J, Feldman JL, Peterson CL, Tan S, Armache JP

PDB-8g57: 
Structure of nucleosome-bound Sirtuin 6 deacetylase
Method: single particle / : Chio US, Rechiche O, Bryll AR, Zhu J, Feldman JL, Peterson CL, Tan S, Armache JP

EMDB-14993: 
Oligomeric structure of SynDLP
Method: single particle / : Gewehr L, Junglas B, Jilly R, Franz J, Wenyu EZ, Weidner T, Bonn M, Sachse C, Schneider D

PDB-7zw6: 
Oligomeric structure of SynDLP
Method: single particle / : Gewehr L, Junglas B, Jilly R, Franz J, Wenyu EZ, Weidner T, Bonn M, Sachse C, Schneider D

EMDB-33891: 
Cryo-EM structure of the human chemerin receptor 1 complex with the C-terminal nonapeptide of chemerin
Method: single particle / : Chen G, Liao Q, Ye RD, Wang J

PDB-7ykd: 
Cryo-EM structure of the human chemerin receptor 1 complex with the C-terminal nonapeptide of chemerin
Method: single particle / : Chen G, Liao Q, Ye RD, Wang J

EMDB-26427: 
Structure of recombinantly assembled A53E alpha-synuclein fibrils
Method: helical / : Zhou K, Zhou H

PDB-7uak: 
Structure of recombinantly assembled A53E alpha-synuclein fibrils
Method: helical / : Zhou K, Zhou H

EMDB-15700: 
Type 3 secretion system needle complex of Shigella flexneri
Method: single particle / : Lunelli M

EMDB-15701: 
Outer membrane secretin pore of the type 3 secretion system of Shigella flexneri
Method: single particle / : Lunelli M

EMDB-15702: 
Inner membrane ring and secretin N0 N1 domains of the type 3 secretion system of Shigella flexneri
Method: single particle / : Lunelli M

EMDB-15703: 
Inner membrane ring of the type 3 secretion system of Shigella flexneri
Method: single particle / : Lunelli M

PDB-8axk: 
Type 3 secretion system export apparatus core, inner rod and needle of Shigella flexneri
Method: single particle / : Lunelli M

PDB-8axl: 
Outer membrane secretin pore of the type 3 secretion system of Shigella flexneri
Method: single particle / : Lunelli M

PDB-8axn: 
Inner membrane ring and secretin N0 N1 domains of the type 3 secretion system of Shigella flexneri
Method: single particle / : Lunelli M
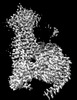
EMDB-32006: 
Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with S1P
Method: single particle / : Yu LY, Gan B, Xiao QJ, Ren RB
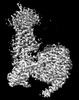
EMDB-32007: 
Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with (S)-FTY720-P
Method: single particle / : Yu LY, Gan B, Xiao QJ, Ren RB
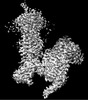
EMDB-32008: 
Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307
Method: single particle / : Yu LY, Gan B, Xiao QJ, Ren RB

EMDB-32009: 
Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307
Method: single particle / : Yu LY, Gan B, Xiao QJ, Ren RB

EMDB-31962: 
The structure of Formyl Peptide Receptor 1 in complex with Gi and peptide agonist fMIFL
Method: single particle / : Wang XK, Chen G, Liao QW, Du Y, Hu HL, Ye DQ

PDB-7vfx: 
The structure of Formyl Peptide Receptor 1 in complex with Gi and peptide agonist fMIFL
Method: single particle / : Wang XK, Chen G, Liao QW, Du Y, Hu HL, Ye DQ

EMDB-26507: 
SARS-CoV-2 spike in complex with Multivalent miniprotein inhibitor FUS231-P24 (2RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26508: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS231-P24 (3RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26509: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (2RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26510: 
SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (3RBDs open)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26511: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-26512: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-7uhb: 
SARS-CoV-2 spike in complex with AHB2-2GS-SB175 (local refinement of the RBD and AHB2)
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
