[English] 日本語
 Yorodumi
Yorodumi- EMDB-15701: Outer membrane secretin pore of the type 3 secretion system of Sh... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Outer membrane secretin pore of the type 3 secretion system of Shigella flexneri | |||||||||
 Map data Map data | Main map sharpened and masked | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | type 3 secretion / secretin / outer membrane ring / shigella / infection / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationtype III protein secretion system complex / type II protein secretion system complex / protein secretion by the type III secretion system / cell outer membrane Similarity search - Function | |||||||||
| Biological species |  Shigella flexneri (bacteria) Shigella flexneri (bacteria) | |||||||||
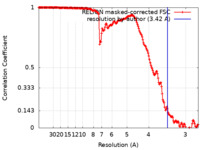
| Method | single particle reconstruction / cryo EM / Resolution: 3.42 Å | |||||||||
 Authors Authors | Lunelli M | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Protein Sci / Year: 2023 Journal: Protein Sci / Year: 2023Title: Integrative structural analysis of the type III secretion system needle complex from Shigella flexneri. Authors: Lara Flacht / Michele Lunelli / Karol Kaszuba / Zhuo Angel Chen / Francis J O' Reilly / Juri Rappsilber / Jan Kosinski / Michael Kolbe /   Abstract: The type III secretion system (T3SS) is a large, transmembrane protein machinery used by various pathogenic gram-negative bacteria to transport virulence factors into the host cell during infection. ...The type III secretion system (T3SS) is a large, transmembrane protein machinery used by various pathogenic gram-negative bacteria to transport virulence factors into the host cell during infection. Understanding the structure of T3SSs is crucial for future developments of therapeutics that could target this system. However, much of the knowledge about the structure of T3SS is available only for Salmonella, and it is unclear how this large assembly is conserved across species. Here, we combined cryo-electron microscopy, cross-linking mass spectrometry, and integrative modeling to determine the structure of the T3SS needle complex from Shigella flexneri. We show that the Shigella T3SS exhibits unique features distinguishing it from other structurally characterized T3SSs. The secretin pore complex adopts a new fold of its C-terminal S domain and the pilotin MxiM[SctG] locates around the outer surface of the pore. The export apparatus structure exhibits a conserved pseudohelical arrangement but includes the N-terminal domain of the SpaS[SctU] subunit, which was not present in any of the previously published virulence-related T3SS structures. Similar to other T3SSs, however, the apparatus is anchored within the needle complex by a network of flexible linkers that either adjust conformation to connect to equivalent patches on the secretin oligomer or bind distinct surface patches at the same height of the export apparatus. The conserved and unique features delineated by our analysis highlight the necessity to analyze T3SS in a species-specific manner, in order to fully understand the underlying molecular mechanisms of these systems. The structure of the type III secretion system from Shigella flexneri delineates conserved and unique features, which could be used for the development of broad-range therapeutics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15701.map.gz emd_15701.map.gz | 24.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15701-v30.xml emd-15701-v30.xml emd-15701.xml emd-15701.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15701_fsc.xml emd_15701_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_15701.png emd_15701.png | 155.3 KB | ||
| Masks |  emd_15701_msk_1.map emd_15701_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15701.cif.gz emd-15701.cif.gz | 5.7 KB | ||
| Others |  emd_15701_additional_1.map.gz emd_15701_additional_1.map.gz emd_15701_half_map_1.map.gz emd_15701_half_map_1.map.gz emd_15701_half_map_2.map.gz emd_15701_half_map_2.map.gz | 330.6 MB 335.2 MB 335.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15701 http://ftp.pdbj.org/pub/emdb/structures/EMD-15701 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15701 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15701 | HTTPS FTP |
-Related structure data
| Related structure data |  8axlMC  8axkC  8axnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15701.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15701.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map sharpened and masked | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38369 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15701_msk_1.map emd_15701_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Main map unsharpened and unmasked
| File | emd_15701_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map unsharpened and unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15701_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_15701_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Needle complex of the type 3 secretion system
| Entire | Name: Needle complex of the type 3 secretion system |
|---|---|
| Components |
|
-Supramolecule #1: Needle complex of the type 3 secretion system
| Supramolecule | Name: Needle complex of the type 3 secretion system / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
-Supramolecule #2: Outer membrane secretin pore of the type 3 secretion system
| Supramolecule | Name: Outer membrane secretin pore of the type 3 secretion system type: complex / ID: 2 / Parent: 1 / Macromolecule list: all Details: Pore formed by the oligomerization of the domains N3, Secretin and S of MxiD |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
-Macromolecule #1: Outer membrane protein MxiD
| Macromolecule | Name: Outer membrane protein MxiD / type: protein_or_peptide / ID: 1 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
| Molecular weight | Theoretical: 63.230414 KDa |
| Sequence | String: MKKFNIKSLT LLIVLLPLIV NANNIDSHLL EQNDIAKYVA QSDTVGSFFE RFSALLNYPI VVSKQAAKKR ISGEFDLSNP EEMLEKLTL LVGLIWYKDG NALYIYDSGE LISKVILLEN ISLNYLIQYL KDANLYDHRY PIRGNISDKT FYISGPPALV E LVANTATL ...String: MKKFNIKSLT LLIVLLPLIV NANNIDSHLL EQNDIAKYVA QSDTVGSFFE RFSALLNYPI VVSKQAAKKR ISGEFDLSNP EEMLEKLTL LVGLIWYKDG NALYIYDSGE LISKVILLEN ISLNYLIQYL KDANLYDHRY PIRGNISDKT FYISGPPALV E LVANTATL LDKQVSSIGT DKVNFGVIKL KNTFVSDRTY NMRGEDIVIP GVATVVERLL NNGKALSNRQ AQNDPMPPFN IT QKVSEDS NDFSFSSVTN SSILEDVSLI AYPETNSILV KGNDQQIQII RDIITQLDVA KRHIELSLWI IDIDKSELNN LGV NWQGTA SFGDSFGASF NMSSSASIST LDGNKFIASV MALNQKKKAN VVSRPVILTQ ENIPAIFDNN RTFYVSLVGE RNSS LEHVT YGTLINVIPR FSSRGQIEMS LTIEDGTGNS QSNYNYNNEN TSVLPEVGRT KISTIARVPQ GKSLLIGGYT HETNS NEII SIPFLSSIPV IGNVFKYKTS NISNIVRVFL IQPREIKESS YYNTAEYKSL ISEREIQKTT QIIPSETTLL EDEKSL VSY LNY UniProtKB: Type 3 secretion system secretin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-6 / Number grids imaged: 1 / Number real images: 5238 / Average exposure time: 1.5 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 101179 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 72.8 |
|---|---|
| Output model |  PDB-8axl: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)