[English] 日本語
 Yorodumi
Yorodumi- EMDB-26509: SARS-CoV-2 spike in complex with multivalent miniprotein inhibito... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (2RBDs open) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.57 Å | |||||||||
 Authors Authors | Park YJ / Seattle Structural Genomics Center for Infectious Disease (SSGCID) / Veesler D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Transl Med / Year: 2022 Journal: Sci Transl Med / Year: 2022Title: Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice. Authors: Andrew C Hunt / James Brett Case / Young-Jun Park / Longxing Cao / Kejia Wu / Alexandra C Walls / Zhuoming Liu / John E Bowen / Hsien-Wei Yeh / Shally Saini / Louisa Helms / Yan Ting Zhao / ...Authors: Andrew C Hunt / James Brett Case / Young-Jun Park / Longxing Cao / Kejia Wu / Alexandra C Walls / Zhuoming Liu / John E Bowen / Hsien-Wei Yeh / Shally Saini / Louisa Helms / Yan Ting Zhao / Tien-Ying Hsiang / Tyler N Starr / Inna Goreshnik / Lisa Kozodoy / Lauren Carter / Rashmi Ravichandran / Lydia B Green / Wadim L Matochko / Christy A Thomson / Bastian Vögeli / Antje Krüger / Laura A VanBlargan / Rita E Chen / Baoling Ying / Adam L Bailey / Natasha M Kafai / Scott E Boyken / Ajasja Ljubetič / Natasha Edman / George Ueda / Cameron M Chow / Max Johnson / Amin Addetia / Mary-Jane Navarro / Nuttada Panpradist / Michael Gale / Benjamin S Freedman / Jesse D Bloom / Hannele Ruohola-Baker / Sean P J Whelan / Lance Stewart / Michael S Diamond / David Veesler / Michael C Jewett / David Baker /    Abstract: New variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continue to arise and prolong the coronavirus disease 2019 (COVID-19) pandemic. Here, we used a cell-free expression ...New variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continue to arise and prolong the coronavirus disease 2019 (COVID-19) pandemic. Here, we used a cell-free expression workflow to rapidly screen and optimize constructs containing multiple computationally designed miniprotein inhibitors of SARS-CoV-2. We found the broadest efficacy was achieved with a homotrimeric version of the 75-residue angiotensin-converting enzyme 2 (ACE2) mimic AHB2 (TRI2-2) designed to geometrically match the trimeric spike architecture. Consistent with the design model, in the cryo-electron microscopy structure TRI2-2 forms a tripod at the apex of the spike protein that engaged all three receptor binding domains simultaneously. TRI2-2 neutralized Omicron (B.1.1.529), Delta (B.1.617.2), and all other variants tested with greater potency than the monoclonal antibodies used clinically for the treatment of COVID-19. TRI2-2 also conferred prophylactic and therapeutic protection against SARS-CoV-2 challenge when administered intranasally in mice. Designed miniprotein receptor mimics geometrically arrayed to match pathogen receptor binding sites could be a widely applicable antiviral therapeutic strategy with advantages over antibodies in greater resistance to viral escape and antigenic drift, and advantages over native receptor traps in lower chances of autoimmune responses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26509.map.gz emd_26509.map.gz | 230 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26509-v30.xml emd-26509-v30.xml emd-26509.xml emd-26509.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26509.png emd_26509.png | 45.1 KB | ||
| Others |  emd_26509_additional_1.map.gz emd_26509_additional_1.map.gz emd_26509_half_map_1.map.gz emd_26509_half_map_1.map.gz emd_26509_half_map_2.map.gz emd_26509_half_map_2.map.gz | 121.6 MB 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26509 http://ftp.pdbj.org/pub/emdb/structures/EMD-26509 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26509 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26509 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26509.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26509.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||
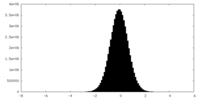
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_26509_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
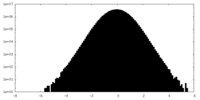
| Density Histograms |
-Half map: #1
| File | emd_26509_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_26509_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike in complex with multivalent miniprotein inhibito...
| Entire | Name: SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (2RBDs open) |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike in complex with multivalent miniprotein inhibito...
| Supramolecule | Name: SARS-CoV-2 spike in complex with multivalent miniprotein inhibitor FUS31-G10 (2RBDs open) type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: SARS-CoV-2 spike
| Supramolecule | Name: SARS-CoV-2 spike / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Multivalent miniprotein inhibitor FUS31-G10
| Supramolecule | Name: Multivalent miniprotein inhibitor FUS31-G10 / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism: Synthetic construct (others) |
| Recombinant expression | Organism:  |
-Macromolecule #1: SARS-CoV-2 Spike
| Macromolecule | Name: SARS-CoV-2 Spike / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSPG SASSVASQSI IAYTMSLGAE NSVAYSNNSI AIPTNFTI S VTTEILPVSM TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGF NFSQILPDPS KPSKRSPIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFG AGPALQIPFP MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STPSALGKLQ DVVNQNAQAL N TLVKQLSS NFGAISSVLN DILSRLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RV DFCGKGY HLMSFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNT FVSGNC DVVIGIVNNT VYDPLQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDL QELGK YEQGSGYIPE APRDGQAYVR KDGEWVLLST FLGRSLEVLF QGPGHHHHHH HHSAWSHPQF EKGGGSGGGG SGGSA WSHP QFEK |
-Macromolecule #2: Multivalent miniprotein inhibitor FUS31-G10
| Macromolecule | Name: Multivalent miniprotein inhibitor FUS31-G10 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: Synthetic construct (others) |
| Recombinant expression | Organism:  |
| Sequence | String: MEKKI SAWS HPQFEK GGG SGGGSGGSAW SHPQFEKGGS GSSG NLDEL HMQMTDLVYE ALHFAKDEEF QKHVFQLFEK ATKAYKNKDR QKLEKVVEEL KELLERLLS GGGSGGGSGG DKENVLQKI YEIMKELERL GHAEASMQVS DLIYEFMKTK DENLLEEAER LLEEVKR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: cryoSPARC ab initio |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.57 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 9733 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)