-Search query
-Search result
Showing 1 - 50 of 70 items for (author: shi & yf)

EMDB-64077: 
Cryo-EM structure of SARS-CoV-2 KP.2 spike RBD in complex with ACE2
Method: single particle / : Jin XH, Sun L

EMDB-64078: 
Cryo-EM structure of SARS-CoV-2 KP.2 spike in complex with ACE2
Method: single particle / : Jin XH, Sun L

EMDB-39865: 
Cryo-EM structure of human GPR4-Gs complex
Method: single particle / : Chen LN, Mao CY, Cheng SZ, Liu YF, Fu YF, Ma XY, Xu P, Ji SY, Wang WW, Shen DD, Zhang HB, Shen QY, Chai R, Zhang M, Yang L, Han F, Cai XJ, Zhang Y

EMDB-39866: 
Cryo-EM structure of human GPR4-Gi complex
Method: single particle / : Chen LN, Zhou H, Xi K, Cheng SZ, Liu YF, Fu YF, Ma XY, Xu P, Ji SY, Wang WW, Shen DD, Zhang HB, Shen QY, Chai R, Zhang M, Yang L, Han F, Mao CY, Cai XJ, Zhang Y

EMDB-63219: 
Cryo-EM structure of human apo inactive GPR4
Method: single particle / : Chen LN, Zhou H, Xi K, Cheng SZ, Liu YF, Fu YF, Ma XY, Xu P, Ji SY, Wang WW, Shen DD, Zhang HB, Shen QY, Chai R, Zhang M, Yang L, Han F, Mao CY, Cai XJ, Zhang Y

EMDB-63220: 
Cryo-EM structure of antagonist-bounded inactive human GPR4
Method: single particle / : Chen LN, Zhou H, Xi K, Cheng SZ, Liu YF, Fu YF, Ma XY, Xu P, Ji SY, Wang WW, Shen DD, Zhang HB, Shen QY, Chai R, Zhang M, Yang L, Han F, Mao CY, Cai XJ, Zhang Y

EMDB-37532: 
Structure of hemagglutinin from Asiatic toad influenza-like virus
Method: single particle / : Zhang D, Xie YF, Chai Y, Shi Y, Liu WJ, Qi JX, Gao GF, Gao F

EMDB-50326: 
Betaglycan in complex with TGF-b1
Method: single particle / : Wieteska L, Coleman JA, Hinck AP

EMDB-50333: 
Betaglycan Orphan Domain (ratBGo) in complex with TGF-b1 and extracellular domain of TGFBRII
Method: single particle / : Wieteska L, Cherepanov P, Punch E, Hinck AP, Hill CS

EMDB-50519: 
Zebrafish Betaglycan Orphan Domain (zfBGo) in complex with TGF-B3 and extracellular domains of TGFBRI and TGFBRII
Method: single particle / : Wieteska L, Coleman JA, Hinck AP

EMDB-50524: 
Zebrafish Betaglycan Orphan Domain (zfBGo) in complex with TGF-b1 and extracellular domain of TGFBRII
Method: single particle / : Wieteska L, Coleman JA, Hinck AP

EMDB-62196: 
MPXV DNA polymerase in complex with primer/5U template DNA
Method: single particle / : Xie YF, Kuai L, Peng Q, Wang Q, Wang H, Li XM, Qi JX, Ding Q, Shi Y, Gao F

EMDB-62197: 
MPXV DNA polymerase in complex with primer/4U template DNA
Method: single particle / : Xie YF, Kuai L, Peng Q, Wang Q, Wang H, Li XM, Qi JX, Ding Q, Shi Y, Gao F

EMDB-62198: 
MPXV DNA polymerase in complex with CDV
Method: single particle / : Xie YF, Kuai L, Peng Q, Wang Q, Wang H, Li XM, Qi JX, Ding Q, Shi Y, Gao F

EMDB-62200: 
MPXV DNA polymerase complex in editing state 2
Method: single particle / : Xie YF, Kuai L, Peng Q, Wang Q, Wang H, Li XM, Qi JX, Ding Q, Shi Y, Gao F

EMDB-62199: 
MPXV DNA polymerase complex in editing state 1
Method: single particle / : Xie YF, Kuai L, Peng Q, Wang Q, Wang H, Li XM, Qi JX, Ding Q, Shi Y, Gao F

EMDB-60516: 
Structure of hemagglutinin from HN/4-10 H3N8 influenza virus G228 mutant complexed with avian receptor analog LSTa
Method: single particle / : Hao TJ, Xie YF, Chai Y, Song H, Gao GF

EMDB-60517: 
Structure of hemagglutinin from HN/4-10 H3N8 influenza virus S228 mutant complexed with avian receptor analog LSTa
Method: single particle / : Hao TJ, Xie YF, Chai Y, Song H, Gao GF

EMDB-60518: 
Structure of hemagglutinin from HN/4-10 H3N8 influenza virus S228 mutant complexed with human receptor analog LSTc
Method: single particle / : Hao TJ, Xie YF, Chai Y, Song H, Gao GF

EMDB-60992: 
Cryo-EM structure of stayfold and nanobody complex from Biortus
Method: single particle / : Shi H, Hu YF, Yang YH, Wang MF, Shi HX

EMDB-39688: 
BA.2.86 RBD protein in complex with ACE2.
Method: single particle / : Wang YJ, Zhang X, Sun L

EMDB-39689: 
Structure of BA.2.86 spike protein in complex with ACE2.
Method: single particle / : Wang YJ, Zang X, Sun L

EMDB-39690: 
Structure of JN.1 RBD protein in complex with ACE2.
Method: single particle / : Wang YJ, Zhang X, Sun L

EMDB-39691: 
The JN.1 spike protein (S) in complex with ACE2.
Method: single particle / : Wang YJ, Zhang X, Sun L

EMDB-60991: 
Cryo-EM structure of GPR158 from Biortus
Method: single particle / : Shi H, Hu YF, Yang YH, Wang MF, Tian FY

EMDB-38157: 
Structure of hemagglutinin from HN/4-10 H3N8 influenza virus G228 mutant complexed with human receptor analog LSTc
Method: single particle / : Hao TJ, Xie YF, Chai Y, Song H, Gao GF

EMDB-61053: 
cryo-EM structure of human cystic fibrosis transmembrane conductance regulator (CFTR) from Biortus
Method: single particle / : Cao S, Shi H, Yang YH, Li JX, Hu YF
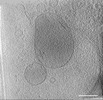
EMDB-39012: 
Representative tomogram of primary glioblastoma stem cell with circular inter-mitochondrial junctions.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39015: 
Representative tomogram of microglia cell with nanotunnel-like structures resembling mitochondrial fission.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39019: 
Representative tomogram of glioblastoma cell with nanotunnel-like structure and inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39021: 
Representative tomogram of normal human astrocyte with nanotunnel-like structure which is an extension of the mitochondrial outer membrane.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39023: 
Representative tomogram of primary glioblastoma differentiated cell with parallel inter-mitochondrial junction.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-39024: 
Representative tomogram of primary glioblastoma stem cell with clustered mitochondria bearing various long-short axis ratios.
Method: electron tomography / : Wang R, Lei H, Wang HX, Qi L, Liu YE, Liu YH, Shi YF, Chen JX, Shen QT

EMDB-29943: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Yang D, Sheng C, Jinpeng S

EMDB-29944: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Jiangqian M, Sheng C, Jinpeng S

EMDB-29945: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Jianqiang M, Jinpeng S

EMDB-29946: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 3 Coupled to G Protein
Method: single particle / : Shenming H, Mengyao X, Lei L, Yang D, Shiyi G, Jinpeng S

EMDB-29935: 
Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein
Method: single particle / : Huang SM, Xiong MY, Liu L, Mu J, Sheng C, Sun J

EMDB-29940: 
Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein
Method: single particle / : Huang SM, Xiong MY, Liu L, Mu J, Sheng C, Sun J

EMDB-35063: 
The complex structure of Omicron BA.4 RBD with BD604, S309, and S304
Method: single particle / : He QW, Xu ZP, Xie YF

EMDB-29016: 
Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane
Method: single particle / : Zhang J, Shi W, Cai YF, Zhu HS, Peng HQ, Voyer J, Volloch SR, Cao H, Mayer ML, Song KK, Xu C, Lu JM, Chen B

EMDB-29017: 
Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane
Method: single particle / : Zhang J, Shi W, Cai YF, Zhu HS, Peng HQ, Voyer J, Volloch SR, Cao H, Mayer ML, Song KK, Xu C, Lu JM, Chen B

EMDB-29018: 
Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane
Method: single particle / : Zhang J, Shi W, Cai YF, Zhu HS, Peng HQ, Voyer J, Volloch SR, Cao H, Mayer ML, Song KK, Xu C, Lu JM, Chen B

EMDB-34731: 
The structure of MPXV polymerase holoenzyme in replicating state
Method: single particle / : Peng Q, Xie YF, Kuai L, Wang H, Qi JX, Gao F, Shi Y

EMDB-25648: 
CryoEM structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab complex with ZM241385
Method: single particle / : Zhang KH, Wu H, Hoppe N, Manglik A, Cheng YF

EMDB-32944: 
The complex structure of Omicron BA.1 RBD with BD604, S309,and S304
Method: single particle / : Huang M, Xie YF

EMDB-32856: 
Cryo-EM structure of SARS-CoV-2 Omicron Spike protein with human ACE2 receptor, C2 state
Method: single particle / : Han WY, Wang YF

EMDB-32857: 
Cryo-EM structure of SARS-CoV-2 Omicron Spike protein with human ACE2 receptor, C3 state
Method: single particle / : Han WY, Wang YF

EMDB-32558: 
Cryo-EM structure of SARS-CoV-2 Omicron spike protein with ACE2, C1 state
Method: single particle / : Han WY, Wang YF

EMDB-32559: 
Cryo-EM structure of Omicron S-ACE2, C2 state
Method: single particle / : Han WY, Wang YF
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
