[English] 日本語
 Yorodumi
Yorodumi- EMDB-25648: CryoEM structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab complex with ZM241385 | |||||||||
 Map data Map data | Structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab with ZM241385 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | A2AAR / GPCR / ADENOSINE RECEPTOR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of norepinephrine secretion / positive regulation of acetylcholine secretion, neurotransmission / negative regulation of alpha-beta T cell activation / positive regulation of circadian sleep/wake cycle, sleep / Adenosine P1 receptors / G protein-coupled adenosine receptor activity / response to purine-containing compound / G protein-coupled adenosine receptor signaling pathway / NGF-independant TRKA activation / Surfactant metabolism ...regulation of norepinephrine secretion / positive regulation of acetylcholine secretion, neurotransmission / negative regulation of alpha-beta T cell activation / positive regulation of circadian sleep/wake cycle, sleep / Adenosine P1 receptors / G protein-coupled adenosine receptor activity / response to purine-containing compound / G protein-coupled adenosine receptor signaling pathway / NGF-independant TRKA activation / Surfactant metabolism / sensory perception / positive regulation of urine volume / synaptic transmission, dopaminergic / type 5 metabotropic glutamate receptor binding / negative regulation of vascular permeability / synaptic transmission, cholinergic / positive regulation of glutamate secretion / intermediate filament / presynaptic active zone / response to caffeine / blood circulation / eating behavior / inhibitory postsynaptic potential / alpha-actinin binding / regulation of calcium ion transport / asymmetric synapse / axolemma / membrane depolarization / phagocytosis / cellular defense response / prepulse inhibition / positive regulation of synaptic transmission, glutamatergic / neuron projection morphogenesis / astrocyte activation / presynaptic modulation of chemical synaptic transmission / response to amphetamine / central nervous system development / positive regulation of long-term synaptic potentiation / positive regulation of synaptic transmission, GABAergic / positive regulation of protein secretion / positive regulation of apoptotic signaling pathway / regulation of mitochondrial membrane potential / synaptic transmission, glutamatergic / excitatory postsynaptic potential / apoptotic signaling pathway / locomotory behavior / electron transport chain / negative regulation of inflammatory response / vasodilation / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / blood coagulation / cell-cell signaling / presynaptic membrane / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / negative regulation of neuron apoptotic process / postsynaptic membrane / calmodulin binding / electron transfer activity / periplasmic space / positive regulation of ERK1 and ERK2 cascade / iron ion binding / response to xenobiotic stimulus / inflammatory response / negative regulation of cell population proliferation / neuronal cell body / heme binding / apoptotic process / dendrite / regulation of DNA-templated transcription / lipid binding / protein-containing complex binding / glutamatergic synapse / enzyme binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Zhang KH / Wu H / Hoppe N / Manglik A / Cheng YF | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Fusion protein strategies for cryo-EM study of G protein-coupled receptors. Authors: Kaihua Zhang / Hao Wu / Nicholas Hoppe / Aashish Manglik / Yifan Cheng /  Abstract: Single particle cryogenic-electron microscopy (cryo-EM) is used extensively to determine structures of activated G protein-coupled receptors (GPCRs) in complex with G proteins or arrestins. However, ...Single particle cryogenic-electron microscopy (cryo-EM) is used extensively to determine structures of activated G protein-coupled receptors (GPCRs) in complex with G proteins or arrestins. However, applying it to GPCRs without signaling proteins remains challenging because most receptors lack structural features in their soluble domains to facilitate image alignment. In GPCR crystallography, inserting a fusion protein between transmembrane helices 5 and 6 is a highly successful strategy for crystallization. Although a similar strategy has the potential to broadly facilitate cryo-EM structure determination of GPCRs alone without signaling protein, the critical determinants that make this approach successful are not yet clear. Here, we address this shortcoming by exploring different fusion protein designs, which lead to structures of antagonist bound A adenosine receptor at 3.4 Å resolution and unliganded Smoothened at 3.7 Å resolution. The fusion strategies explored here are likely applicable to cryo-EM interrogation of other GPCRs and small integral membrane proteins. #1:  Journal: Science / Year: 2012 Journal: Science / Year: 2012Title: Structural Basis for Allosteric Regulation of GPCRs by Sodium Ions Authors: Liu W | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25648.map.gz emd_25648.map.gz | 97.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25648-v30.xml emd-25648-v30.xml emd-25648.xml emd-25648.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25648.png emd_25648.png | 33.5 KB | ||
| Filedesc metadata |  emd-25648.cif.gz emd-25648.cif.gz | 5.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25648 http://ftp.pdbj.org/pub/emdb/structures/EMD-25648 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25648 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25648 | HTTPS FTP |
-Validation report
| Summary document |  emd_25648_validation.pdf.gz emd_25648_validation.pdf.gz | 453.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25648_full_validation.pdf.gz emd_25648_full_validation.pdf.gz | 452.8 KB | Display | |
| Data in XML |  emd_25648_validation.xml.gz emd_25648_validation.xml.gz | 6.4 KB | Display | |
| Data in CIF |  emd_25648_validation.cif.gz emd_25648_validation.cif.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25648 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25648 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25648 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25648 | HTTPS FTP |
-Related structure data
| Related structure data |  7t32MC  8cxoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25648.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25648.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab with ZM241385 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.835 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : A2A adenosine receptor-BRIL/Anti BRIL Fab complex
| Entire | Name: A2A adenosine receptor-BRIL/Anti BRIL Fab complex |
|---|---|
| Components |
|
-Supramolecule #1: A2A adenosine receptor-BRIL/Anti BRIL Fab complex
| Supramolecule | Name: A2A adenosine receptor-BRIL/Anti BRIL Fab complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Adenosine receptor A2a/Soluble cytochrome b562 Fusion Protein
| Macromolecule | Name: Adenosine receptor A2a/Soluble cytochrome b562 Fusion Protein type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.415855 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VYITVELAIA VLAILGNVLV CWAVWLNSNL QNVTNYFVVS LAAADIAVGV LAIPFAITIS TGFCAACHGC LFIACFVLVL TQSSIFSLL AIAIDRYIAI RIPLRYNGLV TGTRAKGIIA ICWVLSFAIG LTPMLGWNNC GQPKEGKNHS QGCGEGQVAC L FEDVVPMN ...String: VYITVELAIA VLAILGNVLV CWAVWLNSNL QNVTNYFVVS LAAADIAVGV LAIPFAITIS TGFCAACHGC LFIACFVLVL TQSSIFSLL AIAIDRYIAI RIPLRYNGLV TGTRAKGIIA ICWVLSFAIG LTPMLGWNNC GQPKEGKNHS QGCGEGQVAC L FEDVVPMN YMVYFNFFAC VLVPLLLMLG VYLRIFLAAR RQLADLEDNW ETLNDNLKVI EKADNAAQVK DALTKMRAAA LD AQKATPP KLEDKSPDSP EMKDFRHGFD ILVGQIDDAL KLANEGKVKE AQAAAEQLKT TRNAYIQKYL ERARSTLQKE VHA AKSLAI IVGLFALCWL PLHIINCFTF FCPDCSHAPL WLMYLAIVLS HTNSVVNPFI YAYRIREFRQ TFRKI UniProtKB: Adenosine receptor A2a, Soluble cytochrome b562, Adenosine receptor A2a |
-Macromolecule #2: 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5...
| Macromolecule | Name: 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol type: ligand / ID: 2 / Number of copies: 1 / Formula: ZMA |
|---|---|
| Molecular weight | Theoretical: 337.336 Da |
| Chemical component information | 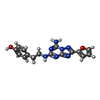 ChemComp-ZMA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 67.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 215946 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















