-Search query
-Search result
Showing 1 - 50 of 105 items for (author: kirk & s)

EMDB-43753: 
Yeast U1 snRNP with humanized U1C Zinc-Finger domain

PDB-8w2o: 
Yeast U1 snRNP with humanized U1C Zinc-Finger domain

EMDB-18207: 
Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide, Glacios map

EMDB-18230: 
Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide
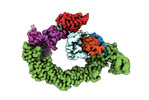
EMDB-18915: 
Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2

PDB-8q7r: 
Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide

PDB-8r5h: 
Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2

EMDB-42804: 
Structure of nucleotide-free Pediculus humanus (Ph) PINK1 dimer

EMDB-42806: 
Structure of AMP-PNP-bound Pediculus humanus (Ph) PINK1 dimer

EMDB-42807: 
Structure of ADP-bound and phosphorylated Pediculus humanus (Ph) PINK1 dimer

PDB-8uyf: 
Structure of nucleotide-free Pediculus humanus (Ph) PINK1 dimer

PDB-8uyh: 
Structure of AMP-PNP-bound Pediculus humanus (Ph) PINK1 dimer

PDB-8uyi: 
Structure of ADP-bound and phosphorylated Pediculus humanus (Ph) PINK1 dimer

EMDB-18586: 
Cryo-EM reconstruction of crosslinked native Bacillus subtilis collided disome binding MutS2 SMR and KOW domains

EMDB-41138: 
CryoEM structure of MFRV-VILP bound to IGF1Rzip

PDB-8tan: 
CryoEM structure of MFRV-VILP bound to IGF1Rzip

EMDB-18585: 
Cryo-EM reconstruction of Bacillus subtilis collided disome binding MutS2 SMR and KOW domains

EMDB-18901: 
Bacillus subtilis MutS2-collided disome complex (stalled 70S)

PDB-8r55: 
Bacillus subtilis MutS2-collided disome complex (collided 70S)

EMDB-18558: 
Bacillus subtilis MutS2-collided disome complex (stalled 70S)

PDB-8qpp: 
Bacillus subtilis MutS2-collided disome complex (stalled 70S)
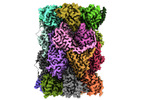
EMDB-29764: 
Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304
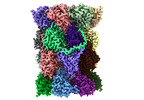
EMDB-29765: 
Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs

EMDB-42148: 
Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304

PDB-8g6e: 
Structure of the Plasmodium falciparum 20S proteasome complexed with inhibitor TDI-8304

PDB-8g6f: 
Structure of the Plasmodium falciparum 20S proteasome beta-6 A117D mutant complexed with inhibitor WLW-vs

PDB-8ud9: 
Structure of human constitutive 20S proteasome complexed with the inhibitor TDI-8304

EMDB-35828: 
Cryo-EPty SPA at CSA of 1.03 mrad

EMDB-35916: 
Cryo-EPty SPA at CSA of 3.26 mrad

EMDB-35917: 
Cryo-EPty SPA at CSA of 4.83 mrad

EMDB-14776: 
Signal peptide mimicry primes Sec61 for client-selective inhibition

PDB-7zl3: 
Signal peptide mimicry primes Sec61 for client-selective inhibition
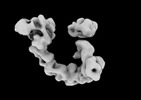
EMDB-29101: 
EGFR:Degrader:VHL:Elongin-B/C:Cul2

EMDB-26306: 
Single-chain LCDV-1 viral insulin-like peptide bound to IGF-1R ectodomain, leucine-zippered form

PDB-7u23: 
Single-chain LCDV-1 viral insulin-like peptide bound to IGF-1R ectodomain, leucine-zippered form

EMDB-26363: 
Head region of insulin receptor ectodomain (A-isoform) bound to the non-insulin agonist IM459

EMDB-26364: 
Head region of insulin receptor ectodomain (A-isoform) bound to the non-insulin agonist IM462

PDB-7u6d: 
Head region of insulin receptor ectodomain (A-isoform) bound to the non-insulin agonist IM459

PDB-7u6e: 
Head region of insulin receptor ectodomain (A-isoform) bound to the non-insulin agonist IM462

EMDB-24791: 
Head region of a complex of IGF-I with the ectodomain of a hybrid insulin receptor / type 1 insulin-like growth factor receptor

EMDB-24927: 
Leg region of a complex of IGF-I with the ectodomain of a hybrid insulin receptor / type 1 insulin-like growth factor receptor

PDB-7s0q: 
Head region of a complex of IGF-I with the ectodomain of a hybrid insulin receptor / type 1 insulin-like growth factor receptor

PDB-7s8v: 
Leg region of a complex of IGF-I with the ectodomain of a hybrid insulin receptor / type 1 insulin-like growth factor receptor

EMDB-13958: 
Structure of the SmrB-bound E. coli disome - collided 70S ribosome

PDB-7qgr: 
Structure of the SmrB-bound E. coli disome - collided 70S ribosome

EMDB-13956: 
Structure of the SmrB-bound E. coli disome - stalled 70S ribosome

PDB-7qgn: 
Structure of the SmrB-bound E. coli disome - stalled 70S ribosome

EMDB-13957: 
Extended H/L (SLPH/SLPL) complex from C. difficile (CD630 strain) fit into R20291 S-layer negative stain map

PDB-7qgq: 
Extended H/L (SLPH/SLPL) complex from C. difficile (CD630 strain) fit into R20291 S-layer negative stain map

EMDB-13952: 
Structure of the collided E. coli disome - VemP-stalled 70S ribosome
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
