+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EPty SPA at CSA of 1.03 mrad | |||||||||||||||
 Map data Map data | Cryo-EPty SPA 3D map reconstructed from 232 particles at convergence angle of 1.03 mrad | |||||||||||||||
 Sample Sample | Rotavirus A != Rotavirus Rotavirus A
| |||||||||||||||
 Keywords Keywords | Complex / VIRUS | |||||||||||||||
| Biological species |  Rotavirus Rotavirus | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 37.2 Å | |||||||||||||||
 Authors Authors | Pei X / Wang P | |||||||||||||||
| Funding support |  China, China,  United Kingdom, European Union, 4 items United Kingdom, European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryogenic electron ptychographic single particle analysis with wide bandwidth information transfer. Authors: Xudong Pei / Liqi Zhou / Chen Huang / Mark Boyce / Judy S Kim / Emanuela Liberti / Yiming Hu / Takeo Sasaki / Peter D Nellist / Peijun Zhang / David I Stuart / Angus I Kirkland / Peng Wang /    Abstract: Advances in cryogenic transmission electron microscopy have revolutionised the determination of many macromolecular structures at atomic or near-atomic resolution. This method is based on ...Advances in cryogenic transmission electron microscopy have revolutionised the determination of many macromolecular structures at atomic or near-atomic resolution. This method is based on conventional defocused phase contrast imaging. However, it has limitations of weaker contrast for small biological molecules embedded in vitreous ice, in comparison with cryo-ptychography, which shows increased contrast. Here we report a single-particle analysis based on the use of ptychographic reconstruction data, demonstrating that three dimensional reconstructions with a wide information transfer bandwidth can be recovered by Fourier domain synthesis. Our work suggests future applications in otherwise challenging single particle analyses, including small macromolecules and heterogeneous or flexible particles. In addition structure determination in situ within cells without the requirement for protein purification and expression may be possible. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35828.map.gz emd_35828.map.gz | 42.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35828-v30.xml emd-35828-v30.xml emd-35828.xml emd-35828.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35828_fsc.xml emd_35828_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_35828.png emd_35828.png | 106.6 KB | ||
| Others |  emd_35828_half_map_1.map.gz emd_35828_half_map_1.map.gz emd_35828_half_map_2.map.gz emd_35828_half_map_2.map.gz | 44.7 MB 44.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35828 http://ftp.pdbj.org/pub/emdb/structures/EMD-35828 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35828 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35828 | HTTPS FTP |
-Validation report
| Summary document |  emd_35828_validation.pdf.gz emd_35828_validation.pdf.gz | 875.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35828_full_validation.pdf.gz emd_35828_full_validation.pdf.gz | 875.2 KB | Display | |
| Data in XML |  emd_35828_validation.xml.gz emd_35828_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_35828_validation.cif.gz emd_35828_validation.cif.gz | 20.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35828 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35828 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35828 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35828 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35828.map.gz / Format: CCP4 / Size: 58.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35828.map.gz / Format: CCP4 / Size: 58.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EPty SPA 3D map reconstructed from 232 particles at convergence angle of 1.03 mrad | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.65 Å | ||||||||||||||||||||||||||||||||||||
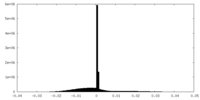
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35828_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
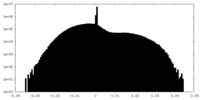
| Density Histograms |
-Half map: #2
| File | emd_35828_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
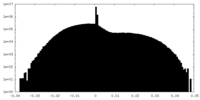
| Density Histograms |
- Sample components
Sample components
-Entire : Rotavirus A
| Entire | Name:  Rotavirus A Rotavirus A |
|---|---|
| Components |
|
-Supramolecule #1: Rotavirus
| Supramolecule | Name: Rotavirus / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 10912 / Sci species name: Rotavirus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: OTHER / Average electron dose: 22.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 14.0 µm / Nominal defocus min: 13.5 µm |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)