[English] 日本語
 Yorodumi
Yorodumi- PDB-8r5h: Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8r5h | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ubiquitin ligation to neosubstrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-VHL-MZ1 with trapped UBE2R2~donor UB-BRD4 BD2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Components Components |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | LIGASE / CUL2 / VHL / ELOBC / BRD4 / MZ1 / PROTAC / Ubiquitin / Ubiquitin Ligase / Monoubiquitylation / NEDD8 / RBX1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of cellular response to hypoxia / cullin-RING-type E3 NEDD8 transferase / NEDD8 transferase activity / RHOBTB3 ATPase cycle / negative regulation of receptor signaling pathway via JAK-STAT / cullin-RING ubiquitin ligase complex / Cul7-RING ubiquitin ligase complex / ubiquitin-dependent protein catabolic process via the C-end degron rule pathway / cellular response to chemical stress / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling ...regulation of cellular response to hypoxia / cullin-RING-type E3 NEDD8 transferase / NEDD8 transferase activity / RHOBTB3 ATPase cycle / negative regulation of receptor signaling pathway via JAK-STAT / cullin-RING ubiquitin ligase complex / Cul7-RING ubiquitin ligase complex / ubiquitin-dependent protein catabolic process via the C-end degron rule pathway / cellular response to chemical stress / Loss of Function of FBXW7 in Cancer and NOTCH1 Signaling / transcription elongation factor activity / target-directed miRNA degradation / elongin complex / positive regulation of protein autoubiquitination / RNA polymerase II transcription initiation surveillance / protein neddylation / Replication of the SARS-CoV-1 genome / NEDD8 ligase activity / VCB complex / negative regulation of response to oxidative stress / Cul5-RING ubiquitin ligase complex / SCF ubiquitin ligase complex / ubiquitin-ubiquitin ligase activity / intracellular membraneless organelle / negative regulation of type I interferon production / E2 ubiquitin-conjugating enzyme / Cul2-RING ubiquitin ligase complex / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / Cul3-RING ubiquitin ligase complex / SUMOylation of ubiquitinylation proteins / negative regulation of mitophagy / Cul4A-RING E3 ubiquitin ligase complex / Cul4-RING E3 ubiquitin ligase complex / Prolactin receptor signaling / Cul4B-RING E3 ubiquitin ligase complex / ubiquitin ligase complex scaffold activity / RNA polymerase II C-terminal domain binding / ubiquitin conjugating enzyme activity / P-TEFb complex binding / negative regulation of DNA damage checkpoint / histone H4 reader activity / negative regulation of transcription elongation by RNA polymerase II / Pausing and recovery of Tat-mediated HIV elongation / Tat-mediated HIV elongation arrest and recovery / cullin family protein binding / HIV elongation arrest and recovery / Pausing and recovery of HIV elongation / host-mediated suppression of viral transcription / positive regulation of G2/M transition of mitotic cell cycle / protein monoubiquitination / negative regulation of signal transduction / positive regulation of T-helper 17 cell lineage commitment / Tat-mediated elongation of the HIV-1 transcript / Formation of HIV-1 elongation complex containing HIV-1 Tat / ubiquitin-like ligase-substrate adaptor activity / Formation of HIV elongation complex in the absence of HIV Tat / protein K48-linked ubiquitination / RNA Polymerase II Transcription Elongation / : / Formation of RNA Pol II elongation complex / Nuclear events stimulated by ALK signaling in cancer / negative regulation of TORC1 signaling / transcription-coupled nucleotide-excision repair / RNA Polymerase II Pre-transcription Events / Maturation of protein E / Maturation of protein E / ER Quality Control Compartment (ERQC) / positive regulation of TORC1 signaling / regulation of cellular response to insulin stimulus / RNA polymerase II CTD heptapeptide repeat kinase activity / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Constitutive Signaling by NOTCH1 HD Domain Mutants / IRAK2 mediated activation of TAK1 complex / Prevention of phagosomal-lysosomal fusion / Alpha-protein kinase 1 signaling pathway / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Endosomal Sorting Complex Required For Transport (ESCRT) / Membrane binding and targetting of GAG proteins / Negative regulation of FLT3 / Regulation of TBK1, IKKε (IKBKE)-mediated activation of IRF3, IRF7 / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / negative regulation of insulin receptor signaling pathway / Regulation of TBK1, IKKε-mediated activation of IRF3, IRF7 upon TLR3 ligation / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / NOTCH2 Activation and Transmission of Signal to the Nucleus / TICAM1,TRAF6-dependent induction of TAK1 complex / TICAM1-dependent activation of IRF3/IRF7 / intrinsic apoptotic signaling pathway / APC/C:Cdc20 mediated degradation of Cyclin B / negative regulation of autophagy / post-translational protein modification / protein serine/threonine kinase binding / Regulation of FZD by ubiquitination / Downregulation of ERBB4 signaling / APC-Cdc20 mediated degradation of Nek2A / p75NTR recruits signalling complexes / InlA-mediated entry of Listeria monocytogenes into host cells Similarity search - Function | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 3.44 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Authors Authors | Liwocha, J. / Prabu, J.R. / Kleiger, G. / Schulman, B.A. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Funding support |  Germany, 1items Germany, 1items
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2024 Journal: Mol Cell / Year: 2024Title: Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting. Authors: Jerry Li / Nicholas Purser / Joanna Liwocha / Daniel C Scott / Holly A Byers / Barbara Steigenberger / Spencer Hill / Ishita Tripathi-Giesgen / Trent Hinkle / Fynn M Hansen / J Rajan Prabu / ...Authors: Jerry Li / Nicholas Purser / Joanna Liwocha / Daniel C Scott / Holly A Byers / Barbara Steigenberger / Spencer Hill / Ishita Tripathi-Giesgen / Trent Hinkle / Fynn M Hansen / J Rajan Prabu / Senthil K Radhakrishnan / Donald S Kirkpatrick / Kurt M Reichermeier / Brenda A Schulman / Gary Kleiger /   Abstract: Cullin-RING ligases (CRLs) ubiquitylate specific substrates selected from other cellular proteins. Substrate discrimination and ubiquitin transferase activity were thought to be strictly separated. ...Cullin-RING ligases (CRLs) ubiquitylate specific substrates selected from other cellular proteins. Substrate discrimination and ubiquitin transferase activity were thought to be strictly separated. Substrates are recognized by substrate receptors, such as Fbox or BCbox proteins. Meanwhile, CRLs employ assorted ubiquitin-carrying enzymes (UCEs, which are a collection of E2 and ARIH-family E3s) specialized for either initial substrate ubiquitylation (priming) or forging poly-ubiquitin chains. We discovered specific human CRL-UCE pairings governing substrate priming. The results reveal pairing of CUL2-based CRLs and UBE2R-family UCEs in cells, essential for efficient PROTAC-induced neo-substrate degradation. Despite UBE2R2's intrinsic programming to catalyze poly-ubiquitylation, CUL2 employs this UCE for geometrically precise PROTAC-dependent ubiquitylation of a neo-substrate and for rapid priming of substrates recruited to diverse receptors. Cryo-EM structures illuminate how CUL2-based CRLs engage UBE2R2 to activate substrate ubiquitylation. Thus, pairing with a specific UCE overcomes E2 catalytic limitations to drive substrate ubiquitylation and targeted protein degradation. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8r5h.cif.gz 8r5h.cif.gz | 276.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8r5h.ent.gz pdb8r5h.ent.gz | 210.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8r5h.json.gz 8r5h.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/r5/8r5h https://data.pdbj.org/pub/pdb/validation_reports/r5/8r5h ftp://data.pdbj.org/pub/pdb/validation_reports/r5/8r5h ftp://data.pdbj.org/pub/pdb/validation_reports/r5/8r5h | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  18915MC  8q7rC C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 8 types, 8 molecules ARCUDFGH
| #1: Protein | Mass: 87098.930 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: CUL2 / Production host: Homo sapiens (human) / Gene: CUL2 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: Q13617 Trichoplusia ni (cabbage looper) / References: UniProt: Q13617 |
|---|---|
| #2: Protein | Mass: 11945.547 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: RBX1, RNF75, ROC1 / Production host: Homo sapiens (human) / Gene: RBX1, RNF75, ROC1 / Production host:  Trichoplusia ni (cabbage looper) / References: UniProt: P62877 Trichoplusia ni (cabbage looper) / References: UniProt: P62877 |
| #3: Protein | Mass: 25853.664 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: UBE2R2, CDC34B, UBC3B / Production host: Homo sapiens (human) / Gene: UBE2R2, CDC34B, UBC3B / Production host:  References: UniProt: Q712K3, E2 ubiquitin-conjugating enzyme |
| #4: Protein | Mass: 8519.778 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: UBC / Production host: Homo sapiens (human) / Gene: UBC / Production host:  |
| #5: Protein | Mass: 12485.135 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: ELOC, TCEB1 / Production host: Homo sapiens (human) / Gene: ELOC, TCEB1 / Production host:  |
| #6: Protein | Mass: 13256.254 Da / Num. of mol.: 1 / Mutation: C356A C357A K368C C391A C429A Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: BRD4, HUNK1 / Production host: Homo sapiens (human) / Gene: BRD4, HUNK1 / Production host:  |
| #7: Protein | Mass: 13147.781 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: ELOB, TCEB2 / Production host: Homo sapiens (human) / Gene: ELOB, TCEB2 / Production host:  |
| #8: Protein | Mass: 18558.162 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: VHL / Production host: Homo sapiens (human) / Gene: VHL / Production host:  |
-Non-polymers , 3 types, 5 molecules 
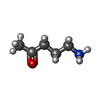


| #9: Chemical | | #10: Chemical | ChemComp-SY8 / | #11: Chemical | ChemComp-759 / ( | |
|---|
-Details
| Has ligand of interest | Y |
|---|---|
| Has protein modification | Y |
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight |
| ||||||||||||||||||||||||
| Source (natural) |
| ||||||||||||||||||||||||
| Source (recombinant) |
| ||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||||||||||
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 130000 X / Nominal defocus max: 2600 nm / Nominal defocus min: 600 nm |
| Image recording | Electron dose: 66 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| EM software | Name: PHENIX / Category: model refinement | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||
| 3D reconstruction | Resolution: 3.44 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 148402 / Symmetry type: POINT | ||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller




 PDBj
PDBj






























