[English] 日本語
 Yorodumi
Yorodumi- EMDB-14776: Signal peptide mimicry primes Sec61 for client-selective inhibition -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Signal peptide mimicry primes Sec61 for client-selective inhibition | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Sec61 Translocon / cryoEM / Membrane Protein / Protein biogenesis | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein transmembrane transporter activity / protein targeting / intracellular protein transport / protein transport / endoplasmic reticulum membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Rehan S / Paavilainen O V | |||||||||
| Funding support |  Finland, 1 items Finland, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: Signal peptide mimicry primes Sec61 for client-selective inhibition. Authors: Shahid Rehan / Dale Tranter / Phillip P Sharp / Gregory B Craven / Eric Lowe / Janet L Anderl / Tony Muchamuel / Vahid Abrishami / Suvi Kuivanen / Nicole A Wenzell / Andy Jennings / ...Authors: Shahid Rehan / Dale Tranter / Phillip P Sharp / Gregory B Craven / Eric Lowe / Janet L Anderl / Tony Muchamuel / Vahid Abrishami / Suvi Kuivanen / Nicole A Wenzell / Andy Jennings / Chakrapani Kalyanaraman / Tomas Strandin / Matti Javanainen / Olli Vapalahti / Matthew P Jacobson / Dustin McMinn / Christopher J Kirk / Juha T Huiskonen / Jack Taunton / Ville O Paavilainen /    Abstract: Preventing the biogenesis of disease-relevant proteins is an attractive therapeutic strategy, but attempts to target essential protein biogenesis factors have been hampered by excessive toxicity. ...Preventing the biogenesis of disease-relevant proteins is an attractive therapeutic strategy, but attempts to target essential protein biogenesis factors have been hampered by excessive toxicity. Here we describe KZR-8445, a cyclic depsipeptide that targets the Sec61 translocon and selectively disrupts secretory and membrane protein biogenesis in a signal peptide-dependent manner. KZR-8445 potently inhibits the secretion of pro-inflammatory cytokines in primary immune cells and is highly efficacious in a mouse model of rheumatoid arthritis. A cryogenic electron microscopy structure reveals that KZR-8445 occupies the fully opened Se61 lateral gate and blocks access to the lumenal plug domain. KZR-8445 binding stabilizes the lateral gate helices in a manner that traps select signal peptides in the Sec61 channel and prevents their movement into the lipid bilayer. Our results establish a framework for the structure-guided discovery of novel therapeutics that selectively modulate Sec61-mediated protein biogenesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14776.map.gz emd_14776.map.gz | 1.6 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14776-v30.xml emd-14776-v30.xml emd-14776.xml emd-14776.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14776_fsc.xml emd_14776_fsc.xml | 28.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_14776.png emd_14776.png | 72.2 KB | ||
| Filedesc metadata |  emd-14776.cif.gz emd-14776.cif.gz | 6.5 KB | ||
| Others |  emd_14776_half_map_1.map.gz emd_14776_half_map_1.map.gz emd_14776_half_map_2.map.gz emd_14776_half_map_2.map.gz | 1.5 GB 1.6 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14776 http://ftp.pdbj.org/pub/emdb/structures/EMD-14776 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14776 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14776 | HTTPS FTP |
-Related structure data
| Related structure data |  7zl3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14776.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14776.map.gz / Format: CCP4 / Size: 1.9 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
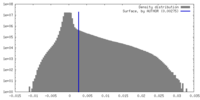
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14776_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
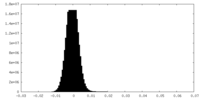
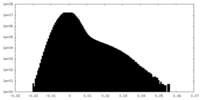
| Density Histograms |
-Half map: #1
| File | emd_14776_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
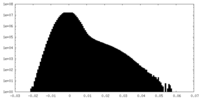
| Density Histograms |
- Sample components
Sample components
-Entire : Protein conducting Sec61 translocon complex
| Entire | Name: Protein conducting Sec61 translocon complex |
|---|---|
| Components |
|
-Supramolecule #1: Protein conducting Sec61 translocon complex
| Supramolecule | Name: Protein conducting Sec61 translocon complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2, #4 |
|---|---|
| Molecular weight | Theoretical: 3.4 MDa |
-Supramolecule #2: Protein conducting Sec61 translocon complex
| Supramolecule | Name: Protein conducting Sec61 translocon complex / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Signal peptide mimic
| Supramolecule | Name: Signal peptide mimic / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: Protein transport protein Sec61 subunit alpha
| Macromolecule | Name: Protein transport protein Sec61 subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.034062 KDa |
| Sequence | String: MGIKFLEVIK PFCVILPEIQ KPERKIQFKE KVLWTAITLF IFLVCCQIPL FGIMSSDSAD PYWMRVILAS NRGTLMALGI SPIVTSGLI MQLLAGAKII EVGDTPKDRA LFNGAQKLFG MTITIGQSIV YVMTGMYGDP SEMGAGVCLL ITIQLFVAGL I VLLLDELL ...String: MGIKFLEVIK PFCVILPEIQ KPERKIQFKE KVLWTAITLF IFLVCCQIPL FGIMSSDSAD PYWMRVILAS NRGTLMALGI SPIVTSGLI MQLLAGAKII EVGDTPKDRA LFNGAQKLFG MTITIGQSIV YVMTGMYGDP SEMGAGVCLL ITIQLFVAGL I VLLLDELL QKGYGLGSGI SLFIATNICE TIVWKAFSPT TVNTGRGMEF EGAIIALFHL LATRTDKVRA LREAFYRQNL PN LMNLIAT IFVFAVVIYF QGFRVDLPIK SARYRGQYNT YPIKLFYTSN IPIILQSALV SNLYVISQML SARFSGNLLV SLL GTWSDT SSGGPARAYP VGGLCHYLSP PESFGSVLED PVHAVVYIVF MLGSCAFFSK TWIEVSGSSA KDVAKQLKEQ QMVM RGHRE TSMVHELNRY IPTAAAFGGL CIGALSVLAD FLGAIGSGTG ILLAVTIIYQ YFEIFVKEQS EVGSMGALLF UniProtKB: Translocon Sec61/SecY plug domain-containing protein |
-Macromolecule #2: Protein transport protein Sec61 subunit gamma
| Macromolecule | Name: Protein transport protein Sec61 subunit gamma / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.752325 KDa |
| Sequence | String: MDQVMQFVEP SRQFVKDSIR LVKRCTKPDR KEFQKIAMAT AIGFAIMGFI GFFVKLIHIP INNIIVGG UniProtKB: Protein transport protein Sec61 subunit gamma |
-Macromolecule #3: Protein transport protein Sec61 subunit beta
| Macromolecule | Name: Protein transport protein Sec61 subunit beta / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.486056 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #4: Cyclic depsipeptide signal peptide mimic
| Macromolecule | Name: Cyclic depsipeptide signal peptide mimic / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 1.101966 KDa |
| Sequence | String: (JMO)(3EG)(MLE)(3EG)(JMX)L |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 0.2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283 K / Instrument: LEICA EM GP | ||||||||||||||||||
| Details | Purified Ribosome-Sec61 complex |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 30294 / Average electron dose: 47.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: OTHER / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)