[English] 日本語
 Yorodumi
Yorodumi- EMDB-26364: Head region of insulin receptor ectodomain (A-isoform) bound to t... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Head region of insulin receptor ectodomain (A-isoform) bound to the non-insulin agonist IM462 | |||||||||
 Map data Map data | Half-map 2 of head region of insulin receptor, leucine-zippered form, bound to insulin receptor peptide IM462 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | insulin receptor / insulin-mimic peptide / insulin receptor agonist / HORMONE-SIGNALING PROTEIN-AGONIST complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of glycogen catabolic process / positive regulation of nitric oxide mediated signal transduction / negative regulation of fatty acid metabolic process / negative regulation of feeding behavior / Signaling by Insulin receptor / IRS activation / regulation of protein secretion / Insulin processing / positive regulation of peptide hormone secretion / positive regulation of respiratory burst ...negative regulation of glycogen catabolic process / positive regulation of nitric oxide mediated signal transduction / negative regulation of fatty acid metabolic process / negative regulation of feeding behavior / Signaling by Insulin receptor / IRS activation / regulation of protein secretion / Insulin processing / positive regulation of peptide hormone secretion / positive regulation of respiratory burst / negative regulation of acute inflammatory response / Regulation of gene expression in beta cells / alpha-beta T cell activation / positive regulation of dendritic spine maintenance / Synthesis, secretion, and deacylation of Ghrelin / activation of protein kinase B activity / negative regulation of protein secretion / negative regulation of gluconeogenesis / positive regulation of glycogen biosynthetic process / fatty acid homeostasis / Signal attenuation / positive regulation of insulin receptor signaling pathway / negative regulation of respiratory burst involved in inflammatory response / FOXO-mediated transcription of oxidative stress, metabolic and neuronal genes / negative regulation of lipid catabolic process / positive regulation of lipid biosynthetic process / negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway / regulation of protein localization to plasma membrane / nitric oxide-cGMP-mediated signaling / transport vesicle / COPI-mediated anterograde transport / positive regulation of nitric-oxide synthase activity / Insulin receptor recycling / negative regulation of reactive oxygen species biosynthetic process / positive regulation of brown fat cell differentiation / insulin-like growth factor receptor binding / NPAS4 regulates expression of target genes / neuron projection maintenance / endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of mitotic nuclear division / Insulin receptor signalling cascade / positive regulation of glycolytic process / positive regulation of cytokine production / endosome lumen / positive regulation of long-term synaptic potentiation / acute-phase response / positive regulation of protein secretion / positive regulation of D-glucose import across plasma membrane / insulin receptor binding / positive regulation of cell differentiation / Regulation of insulin secretion / wound healing / positive regulation of neuron projection development / receptor protein-tyrosine kinase / hormone activity / regulation of synaptic plasticity / negative regulation of protein catabolic process / positive regulation of protein localization to nucleus / Golgi lumen / vasodilation / cognition / glucose metabolic process / insulin receptor signaling pathway / cell-cell signaling / glucose homeostasis / regulation of protein localization / PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling / positive regulation of cell growth / protease binding / secretory granule lumen / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / positive regulation of canonical NF-kappaB signal transduction / positive regulation of MAPK cascade / positive regulation of cell migration / G protein-coupled receptor signaling pathway / endoplasmic reticulum lumen / Amyloid fiber formation / Golgi membrane / negative regulation of gene expression / positive regulation of cell population proliferation / positive regulation of gene expression / regulation of DNA-templated transcription / extracellular space / extracellular region / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Kirk NS / Lawrence MC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Activation of the human insulin receptor by non-insulin-related peptides. Authors: Nicholas S Kirk / Qi Chen / Yingzhe Ginger Wu / Anastasia L Asante / Haitao Hu / Juan F Espinosa / Francisco Martínez-Olid / Mai B Margetts / Faiz A Mohammed / Vladislav V Kiselyov / David ...Authors: Nicholas S Kirk / Qi Chen / Yingzhe Ginger Wu / Anastasia L Asante / Haitao Hu / Juan F Espinosa / Francisco Martínez-Olid / Mai B Margetts / Faiz A Mohammed / Vladislav V Kiselyov / David G Barrett / Michael C Lawrence /    Abstract: The human insulin receptor signalling system plays a critical role in glucose homeostasis. Insulin binding brings about extensive conformational change in the receptor extracellular region that in ...The human insulin receptor signalling system plays a critical role in glucose homeostasis. Insulin binding brings about extensive conformational change in the receptor extracellular region that in turn effects trans-activation of the intracellular tyrosine kinase domains and downstream signalling. Of particular therapeutic interest is whether insulin receptor signalling can be replicated by molecules other than insulin. Here, we present single-particle cryoEM structures that show how a 33-mer polypeptide unrelated to insulin can cross-link two sites on the receptor surface and direct the receptor into a signalling-active conformation. The 33-mer polypeptide engages the receptor by two helical binding motifs that are each potentially mimicable by small molecules. The resultant conformation of the receptor is distinct from-but related to-those in extant three-dimensional structures of the insulin-complexed receptor. Our findings thus illuminate unexplored pathways for controlling the signalling of the insulin receptor as well as opportunities for development of insulin mimetics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26364.map.gz emd_26364.map.gz | 5.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26364-v30.xml emd-26364-v30.xml emd-26364.xml emd-26364.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26364.png emd_26364.png | 75.1 KB | ||
| Filedesc metadata |  emd-26364.cif.gz emd-26364.cif.gz | 6.9 KB | ||
| Others |  emd_26364_half_map_1.map.gz emd_26364_half_map_1.map.gz emd_26364_half_map_2.map.gz emd_26364_half_map_2.map.gz | 71.3 MB 71.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26364 http://ftp.pdbj.org/pub/emdb/structures/EMD-26364 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26364 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26364 | HTTPS FTP |
-Related structure data
| Related structure data |  7u6eMC  7u6dC  8di2C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26364.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26364.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
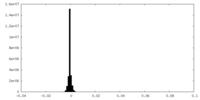
| Annotation | Half-map 2 of head region of insulin receptor, leucine-zippered form, bound to insulin receptor peptide IM462 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.41333 Å | ||||||||||||||||||||||||||||||||||||
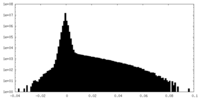
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-map 1 of head region of insulin receptor,...
| File | emd_26364_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
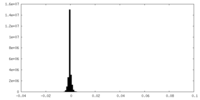
| Annotation | Half-map 1 of head region of insulin receptor, leucine-zippered form, bound to insulin receptor peptide IM462 | ||||||||||||
| Projections & Slices |
| ||||||||||||
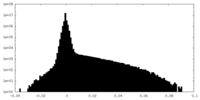
| Density Histograms |
-Half map: Half-map 1 of head region of insulin receptor,...
| File | emd_26364_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 of head region of insulin receptor, leucine-zippered form, bound to insulin receptor peptide IM462 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of IM462 with insulin and Fv 83-7 bound to insulin recept...
| Entire | Name: Complex of IM462 with insulin and Fv 83-7 bound to insulin receptor, delta beta, leucine zippered form |
|---|---|
| Components |
|
-Supramolecule #1: Complex of IM462 with insulin and Fv 83-7 bound to insulin recept...
| Supramolecule | Name: Complex of IM462 with insulin and Fv 83-7 bound to insulin receptor, delta beta, leucine zippered form type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Insulin A chain
| Macromolecule | Name: Insulin A chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.383698 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GIVEQCCTSI CSLYQLENYC N UniProtKB: Insulin |
-Macromolecule #2: Insulin B chain
| Macromolecule | Name: Insulin B chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 3.433953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FVNQHLCGSH LVEALYLVCG ERGFFYTPKT UniProtKB: Insulin |
-Macromolecule #3: Isoform Short of Insulin receptor
| Macromolecule | Name: Isoform Short of Insulin receptor / type: protein_or_peptide / ID: 3 / Details: delta beta construct, leucine-zippered form / Number of copies: 2 / Enantiomer: LEVO / EC number: receptor protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 106.728211 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HLYPGEVCPG MDIRNNLTRL HELENCSVIE GHLQILLMFK TRPEDFRDLS FPKLIMITDY LLLFRVYGLE SLKDLFPNLT VIRGSRLFF NYALVIFEMV HLKELGLYNL MNITRGSVRI EKNNELCYLA TIDWSRILDS VEDNHIVLNK DDNEECGDIC P GTAKGKTN ...String: HLYPGEVCPG MDIRNNLTRL HELENCSVIE GHLQILLMFK TRPEDFRDLS FPKLIMITDY LLLFRVYGLE SLKDLFPNLT VIRGSRLFF NYALVIFEMV HLKELGLYNL MNITRGSVRI EKNNELCYLA TIDWSRILDS VEDNHIVLNK DDNEECGDIC P GTAKGKTN CPATVINGQF VERCWTHSHC QKVCPTICKS HGCTAEGLCC HSECLGNCSQ PDDPTKCVAC RNFYLDGRCV ET CPPPYYH FQDWRCVNFS FCQDLHHKCK NSRRQGCHQY VIHNNKCIPE CPSGYTMNSS NLLCTPCLGP CPKVCHLLEG EKT IDSVTS AQELRGCTVI NGSLIINIRG GNNLAAELEA NLGLIEEISG YLKIRRSYAL VSLSFFRKLR LIRGETLEIG NYSF YALDN QNLRQLWDWS KHNLTTTQGK LFFHYNPKLC LSEIHKMEEV SGTKGRQERN DIALKTNGDK ASCENELLKF SYIRT SFDK ILLRWEPYWP PDFRDLLGFM LFYKEAPYQN VTEFDGQDAC GSNSWTVVDI DPPLRSNDPK SQNHPGWLMR GLKPWT QYA IFVKTLVTFS DERRTYGAKS DIIYVQTDAT NPSVPLDPIS VSNSSSQIIL KWKPPSDPNG NITHYLVFWE RQAEDSE LF ELDYCLKGLK LPSRTWSPPF ESEDSQKHNQ SEYEDSAGEC CSCPKTDSQI LKELEESSFR KTFEDYLHNV VFVPRPSR K RRSLGDVGNA GNNEEHRPFE KVVNKESLVI SGLRHFTGYR IELQACNQDT PEERCSVAAY VSARTMPEAK ADDIVGPVT HEIFENNVVH LMWQEPKEPN GLIVLYEVSY RRYGDEELHL CVSRKHFALE RGCRLRGLSP GNYSVRIRAT SLAGNGSWTE PTYFYVTDY LDVPSNIARM KQLEDKVEEL LSKNYHLENE VARLKKLVGE R UniProtKB: Isoform Short of Insulin receptor |
-Macromolecule #4: IM462
| Macromolecule | Name: IM462 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 2.644862 KDa |
| Sequence | String: (HY1)SLEEEWAQI ECEVYGRCPP SES |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)