+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
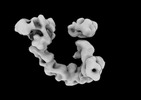
| Title | EGFR:Degrader:VHL:Elongin-B/C:Cul2 | |||||||||
 Map data Map data | EGFR:Degrader:VHL:Elongin-B/C:Cul2 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 6.2 Å | |||||||||
 Authors Authors | Rosenberg SC | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Chem Biol / Year: 2023 Journal: Cell Chem Biol / Year: 2023Title: Ternary complex dissociation kinetics contribute to mutant-selective EGFR degradation. Authors: Scott C Rosenberg / Frances Shanahan / Sayumi Yamazoe / Marc Kschonsak / Yi J Zeng / James Lee / Emile Plise / Ivana Yen / Christopher M Rose / John G Quinn / Lewis J Gazzard / Benjamin T ...Authors: Scott C Rosenberg / Frances Shanahan / Sayumi Yamazoe / Marc Kschonsak / Yi J Zeng / James Lee / Emile Plise / Ivana Yen / Christopher M Rose / John G Quinn / Lewis J Gazzard / Benjamin T Walters / Donald S Kirkpatrick / Steven T Staben / Scott A Foster / Shiva Malek /  Abstract: Targeted degradation of proteins by chimeric heterobifunctional degraders has emerged as a major drug discovery paradigm. Despite the increased interest in this approach, the criteria dictating ...Targeted degradation of proteins by chimeric heterobifunctional degraders has emerged as a major drug discovery paradigm. Despite the increased interest in this approach, the criteria dictating target protein degradation by a degrader remain poorly understood, and potent target engagement by a degrader does not strongly correlate with target degradation. In this study, we present the biochemical characterization of an epidermal growth factor receptor (EGFR) degrader that potently binds both wild-type and mutant EGFR, but only degrades EGFR mutant variants. Mechanistic studies reveal that ternary complex half-life strongly correlates with processive ubiquitination with purified components and mutant-selective degradation in cells. We present cryoelectron microscopy and hydrogen-deuterium exchange mass spectroscopy data on wild-type and mutant EGFR ternary complexes, which demonstrate that potent target degradation can be achieved in the absence of stable compound-induced protein-protein interactions. These results highlight the importance of considering target conformation during degrader development as well as leveraging heterobifunctional ligand binding kinetics to achieve robust target degradation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29101.map.gz emd_29101.map.gz | 28.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29101-v30.xml emd-29101-v30.xml emd-29101.xml emd-29101.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29101.png emd_29101.png | 33.5 KB | ||
| Others |  emd_29101_half_map_1.map.gz emd_29101_half_map_1.map.gz emd_29101_half_map_2.map.gz emd_29101_half_map_2.map.gz | 3.9 MB 3.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29101 http://ftp.pdbj.org/pub/emdb/structures/EMD-29101 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29101 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29101 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29101.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29101.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EGFR:Degrader:VHL:Elongin-B/C:Cul2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_29101_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
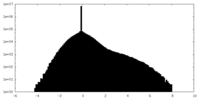
| Density Histograms |
-Half map: Half Map 2
| File | emd_29101_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
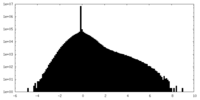
| Density Histograms |
- Sample components
Sample components
-Entire : Degrader induced complex between EGFR and Cul2:Rbx1:VHL:Elongin-B/C
| Entire | Name: Degrader induced complex between EGFR and Cul2:Rbx1:VHL:Elongin-B/C |
|---|---|
| Components |
|
-Supramolecule #1: Degrader induced complex between EGFR and Cul2:Rbx1:VHL:Elongin-B/C
| Supramolecule | Name: Degrader induced complex between EGFR and Cul2:Rbx1:VHL:Elongin-B/C type: complex / ID: 1 / Chimera: Yes / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 200 mM NaCl, 20 mM Tris7.5, 1 mM TCEP |
| Grid | Model: C-flat-1.2/1.3 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number real images: 1848 / Average exposure time: 20.0 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.0015 µm / Nominal defocus min: 0.0005 µm |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 6.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 55053 |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)