-Search query
-Search result
Showing all 42 items for (author: guerra & m)

EMDB-18182: 
Closed conformation of the g-tubulin ring complex nucleating microtubules

EMDB-18181: 
Early closed conformation of the g-tubulin ring complex

PDB-8q62: 
Early closed conformation of the g-tubulin ring complex

EMDB-28850: 
SARS-CoV-2 Gamma 6P Mut7 S + COVA309-3 Fab

EMDB-28851: 
SARS-CoV-2 Gamma 6P Mut7 S + COVA309-10 Fab

EMDB-28852: 
SARS-CoV-2 Omicron 6P S + COVA309-35 Fab
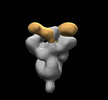
EMDB-28853: 
SARS-CoV-2 Gamma 6P Mut7 + S COVA309-38 Fab

EMDB-27177: 
sd1.040 Fab in complex with SARS-CoV-2 Spike 2P glycoprotein

PDB-8d48: 
sd1.040 Fab in complex with SARS-CoV-2 Spike 2P glycoprotein

EMDB-25412: 
Structure of the human COQ7:COQ9 complex by single-particle electron cryo-microscopy, unliganded state

EMDB-25413: 
Structure of the NADH-bound human COQ7:COQ9 complex by single-particle electron cryo-microscopy

PDB-7ssp: 
Structure of the human COQ7:COQ9 complex by single-particle electron cryo-microscopy, unliganded state

PDB-7sss: 
Structure of the NADH-bound human COQ7:COQ9 complex by single-particle electron cryo-microscopy
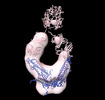
EMDB-26217: 
Negative stain EM map of COVA1-07 mAb bound to the S2 domain of SARS-CoV-2 S
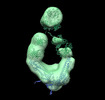
EMDB-26218: 
Negative stain EM map of COVA2-14 mAb bound to the S2 domain of SARS-CoV-2 S

EMDB-26219: 
Negative stain EM map of COVA2-18 mAb bound to the S2 domain of SARS-CoV-2 S
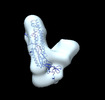
EMDB-26220: 
Negative stain EM map of the S2 domain of SARS-CoV-2 S

EMDB-13482: 
Half-vault structure
PDB-7pky: 
Half-vault structure

EMDB-13478: 
Vault structure in primmed conformation

EMDB-13483: 
Vault structure in committed conformation

PDB-7pkr: 
Vault structure in primmed conformation

PDB-7pkz: 
Vault structure in committed conformation

PDB-7ko8: 
Cryo-EM structure of the mature and infective Mayaro virus

EMDB-22961: 
Cryo-EM structure of the mature and infective Mayaro virus

EMDB-23502: 
Cryo-EM structure of the Pre3-1 20S proteasome core particle

EMDB-23503: 
Cryo-EM structure of Pre-15S proteasome core particle assembly intermediate purified from Pre3-1 proteasome mutant (G34D)

EMDB-23508: 
Cryo-EM structure of 13S proteasome core particle assembly intermediate purified from Pre3-1 proteasome mutant (G34D)

PDB-7ls5: 
Cryo-EM structure of the Pre3-1 20S proteasome core particle

PDB-7ls6: 
Cryo-EM structure of Pre-15S proteasome core particle assembly intermediate purified from Pre3-1 proteasome mutant (G34D)

PDB-7lsx: 
Cryo-EM structure of 13S proteasome core particle assembly intermediate purified from Pre3-1 proteasome mutant (G34D)

EMDB-11236: 
Reconstruction of Tula virus surface glycoprotein lattice

PDB-6zjm: 
Atomic model of Andes virus glycoprotein spike tetramer generated by fitting into a Tula virus reconstruction
EMDB-22061: 
negative stain EM map of SARS-CoV-2 spike in complex with COVA2-15 Fab

EMDB-22062: 
negative stain EM map of SARS-CoV-2 spike in complex with COVA1-22

EMDB-22063: 
negative stain EM map of SARS-CoV-2 spike in complex with COVA2-07

EMDB-22064: 
negative stain EM map of SARS-CoV-2 spike in complex with COVA2-39 Fab

EMDB-22065: 
negative stain EM map of SARS-CoV-2 spike in complex with COVA2-04 Fab

EMDB-22066: 
negative stain EM map of SARS-CoV-2 spike in complex with COVA1-12 Fab

EMDB-0340: 
EM structure of the DNA wrapping in bacterial open transcription initiation complex

EMDB-4867: 
Sub-tomogram averaging of Tula virus glycoprotein spike

EMDB-2101: 
Electron tomography of forming Hepatitis C virus-like particles in insect cells.
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
