+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Gamma 6P Mut7 S + COVA309-3 Fab | |||||||||
 Map data Map data | Gamma 6P Mut7 COVA309-3 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | coronavirus / SARS-CoV-2 / COVID / structural protein / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Torres JL / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: iScience / Year: 2023 Journal: iScience / Year: 2023Title: Broad SARS-CoV-2 neutralization by monoclonal and bispecific antibodies derived from a Gamma-infected individual. Authors: Denise Guerra / Tim Beaumont / Laura Radić / Gius Kerster / Karlijn van der Straten / Meng Yuan / Jonathan L Torres / Wen-Hsin Lee / Hejun Liu / Meliawati Poniman / Ilja Bontjer / Judith A ...Authors: Denise Guerra / Tim Beaumont / Laura Radić / Gius Kerster / Karlijn van der Straten / Meng Yuan / Jonathan L Torres / Wen-Hsin Lee / Hejun Liu / Meliawati Poniman / Ilja Bontjer / Judith A Burger / Mathieu Claireaux / Tom G Caniels / Jonne L Snitselaar / Tom P L Bijl / Sabine Kruijer / Gabriel Ozorowski / David Gideonse / Kwinten Sliepen / Andrew B Ward / Dirk Eggink / Godelieve J de Bree / Ian A Wilson / Rogier W Sanders / Marit J van Gils /   Abstract: The pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has remained a medical threat due to the evolution of multiple variants that acquire resistance to vaccines and ...The pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has remained a medical threat due to the evolution of multiple variants that acquire resistance to vaccines and prior infection. Therefore, it is imperative to discover monoclonal antibodies (mAbs) that neutralize a broad range of SARS-CoV-2 variants. A stabilized spike glycoprotein was used to enrich antigen-specific B cells from an individual with a primary Gamma variant infection. Five mAbs selected from those B cells showed considerable neutralizing potency against multiple variants, with COVA309-35 being the most potent against the autologous virus, as well as Omicron BA.1 and BA.2, and COVA309-22 having binding and neutralization activity against Omicron BA.4/5, BQ.1.1, and XBB.1. When combining the COVA309 mAbs as cocktails or bispecific antibodies, the breadth and potency were improved. In addition, the mechanism of cross-neutralization of the COVA309 mAbs was elucidated by structural analysis. Altogether these data indicate that a Gamma-infected individual can develop broadly neutralizing antibodies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28850.map.gz emd_28850.map.gz | 49.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28850-v30.xml emd-28850-v30.xml emd-28850.xml emd-28850.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28850.png emd_28850.png | 23.8 KB | ||
| Filedesc metadata |  emd-28850.cif.gz emd-28850.cif.gz | 3.9 KB | ||
| Others |  emd_28850_half_map_1.map.gz emd_28850_half_map_1.map.gz emd_28850_half_map_2.map.gz emd_28850_half_map_2.map.gz | 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28850 http://ftp.pdbj.org/pub/emdb/structures/EMD-28850 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28850 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28850 | HTTPS FTP |
-Validation report
| Summary document |  emd_28850_validation.pdf.gz emd_28850_validation.pdf.gz | 658.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28850_full_validation.pdf.gz emd_28850_full_validation.pdf.gz | 658.5 KB | Display | |
| Data in XML |  emd_28850_validation.xml.gz emd_28850_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_28850_validation.cif.gz emd_28850_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28850 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28850 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28850 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28850 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28850.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28850.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Gamma 6P Mut7 COVA309-3 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.06 Å | ||||||||||||||||||||||||||||||||||||
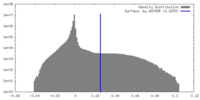
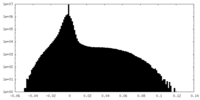
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Gamma 6P Mut7 COVA309-3
| File | emd_28850_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Gamma 6P Mut7 COVA309-3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
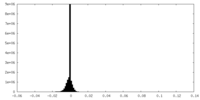
| Density Histograms |
-Half map: Gamma 6P Mut7 COVA309-3
| File | emd_28850_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Gamma 6P Mut7 COVA309-3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
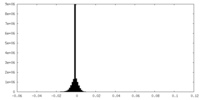
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Gamma 6P Mut7 S + COVA309-3 Fab
| Entire | Name: SARS-CoV-2 Gamma 6P Mut7 S + COVA309-3 Fab |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Gamma 6P Mut7 S + COVA309-3 Fab
| Supramolecule | Name: SARS-CoV-2 Gamma 6P Mut7 S + COVA309-3 Fab / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Staining | Type: NEGATIVE / Material: 2% Uranyl Formate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.5 CUT-OFF / Number images used: 19302 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)