[English] 日本語
 Yorodumi
Yorodumi- EMDB-0340: EM structure of the DNA wrapping in bacterial open transcription ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0340 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EM structure of the DNA wrapping in bacterial open transcription initiation complex | |||||||||
 Map data Map data | EM map of DNA wrapping in bacterial transcription initiation complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA wrapping / bacterial transcription initiation complex / transmission electron microscopy / single particle analysis. / TRANSCRIPTION / transcription-dna complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsigma factor antagonist complex / RNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / sigma factor activity / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility ...sigma factor antagonist complex / RNA polymerase complex / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / sigma factor activity / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility / nitrate assimilation / regulation of DNA-templated transcription elongation / transcription elongation factor complex / transcription antitermination / DNA-directed RNA polymerase complex / cell motility / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / response to heat / protein-containing complex assembly / intracellular iron ion homeostasis / protein dimerization activity / transcription cis-regulatory region binding / response to antibiotic / negative regulation of DNA-templated transcription / regulation of DNA-templated transcription / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |    Enterobacteria phage lambda (virus) Enterobacteria phage lambda (virus) | |||||||||
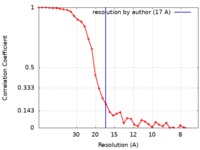
| Method | single particle reconstruction / negative staining / Resolution: 17.0 Å | |||||||||
 Authors Authors | Florez-Ariza A / Cassago A / de Oliveira PSL / Guerra DG | |||||||||
 Citation Citation |  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: Interactions of Upstream and Downstream Promoter Regions with RNA Polymerase are Energetically Coupled and a Target of Regulation in Transcription Initiation Authors: Sosa R / Florez-Ariza A / Diaz-Celis C / Onoa B / Cassago A / de Oliveira PSL / Portugal RV / Guerra DG / Bustamante C | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0340.map.gz emd_0340.map.gz | 958.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0340-v30.xml emd-0340-v30.xml emd-0340.xml emd-0340.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0340_fsc.xml emd_0340_fsc.xml | 3.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_0340.png emd_0340.png | 202.5 KB | ||
| Filedesc metadata |  emd-0340.cif.gz emd-0340.cif.gz | 8.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0340 http://ftp.pdbj.org/pub/emdb/structures/EMD-0340 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0340 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0340 | HTTPS FTP |
-Related structure data
| Related structure data |  6n4cMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0340.map.gz / Format: CCP4 / Size: 2.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0340.map.gz / Format: CCP4 / Size: 2.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map of DNA wrapping in bacterial transcription initiation complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.56 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : E. coli RNAP-DNA wrapped open complex
| Entire | Name: E. coli RNAP-DNA wrapped open complex |
|---|---|
| Components |
|
-Supramolecule #1: E. coli RNAP-DNA wrapped open complex
| Supramolecule | Name: E. coli RNAP-DNA wrapped open complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Wrapped transcription initiation open complex assembled between E. coli RNAP-sigma 70 holoenzyme and lambda PR wild-type promoter (+18 to -76) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: RNA polymerase sigma factor RpoD
| Macromolecule | Name: RNA polymerase sigma factor RpoD / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.322277 KDa |
| Sequence | String: QLKLLVTRGK EQGYLTYAEV NDHLPEDIVD SDQIEDIIQM INDMGIQVME EAPDADDLML AENTADEDAA EAAAQVLSSV ESEIGRTTD PVRMYMREMG TVELLTREGE IDIAKRIEDG INQVQCSVAE YPEAITYLLE QYDRVEAEEA RLSDLITGFV D PNAENSID ...String: QLKLLVTRGK EQGYLTYAEV NDHLPEDIVD SDQIEDIIQM INDMGIQVME EAPDADDLML AENTADEDAA EAAAQVLSSV ESEIGRTTD PVRMYMREMG TVELLTREGE IDIAKRIEDG INQVQCSVAE YPEAITYLLE QYDRVEAEEA RLSDLITGFV D PNAENSID PELAREKFAE LRAQYVVTRD TIKAKGRSHA TAQEEILKLS EVFKQFRLVP KQFDYLVNSM RVMMDRVRTQ ER LIMKLCV EQCKMPKKNF ITLFTGNETS DTWFNAAIAM NKPWSEKLHD VSEEVHRALQ KLQQIEEETG LTIEQVKDIN RRM SIGEAK ARRAKKEMVE ANLRLVISIA KKYTNRGLQF LDLIQEGNIG LMKAVDKFEY RRGYKFSTYA TWWIRQAITR SIAD QARTI RIPVHMIETI NKLNRISRQM LQEMGREPTP EELAERMLMP EDKIRKVLKI SMETPIGDDE DSHLGIDFIE DTTSA TTES LRAATHDVLA GLTAREAKVL RMRFGIDMNT DYTLEEVGKQ FDVTRERIRQ IEAKALRKLR HPSRSEVLRS FLDD UniProtKB: RNA polymerase sigma factor RpoD |
-Macromolecule #2: DNA-directed RNA polymerase subunit beta
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 150.689672 KDa |
| Sequence | String: VYSYTEKKRI RKDFGKRPQV LDVPYLLSIQ LDSFQKFIEQ DPEGQYGLEA AFRSVFPIQS YSGNSELQYV SYRLGEPVFD VQECQIRGV TYSAPLRVKL RLVIYEREAP EGTVKDIKEQ EVYMGEIPLM TDNGTFVING TERVIVSQLH RSPGVFFDSD K GKTHSSGK ...String: VYSYTEKKRI RKDFGKRPQV LDVPYLLSIQ LDSFQKFIEQ DPEGQYGLEA AFRSVFPIQS YSGNSELQYV SYRLGEPVFD VQECQIRGV TYSAPLRVKL RLVIYEREAP EGTVKDIKEQ EVYMGEIPLM TDNGTFVING TERVIVSQLH RSPGVFFDSD K GKTHSSGK VLYNARIIPY RGSWLDFEFD PKDNLFVRID RRRKLPATII LRALNYTTEQ ILDLFFEKVI FEIRDNKLQM EL VPERLRG ETASFDIEAN GKVYVEKGRR ITARHIRQLE KDDVKLIEVP VEYIAGKVVA KDYIDESTGE LICAANMELS LDL LAKLSQ SGHKRIETLF TNDLDHGPYI SETLRVDPTN DRLSALVEIY RMMRPGEPPT REAAESLFEN LFFSEDRYDL SAVG RMKFN RSLLREEIEG SGILSKDDII DVMKKLIDIR NGKGEVDDID HLGNRRIRSV GEMAENQFRV GLVRVERAVK ERLSL GDLD TLMPQDMINA KPISAAVKEF FGSSQLSQFM DQNNPLSEIT HKRRISALGP GGLTRERAGF EVRDVHPTHY GRVCPI ETP EGPNIGLINS LSVYAQTNEY GFLETPYRKV TDGVVTDEIH YLSAIEEGNY VIAQANSNLD EEGHFVEDLV TCRSKGE SS LFSRDQVDYM DVSTQQVVSV GASLIPFLEH DDANRALMGA NMQRQAVPTL RADKPLVGTG MERAVAVDSG VTAVAKRG G VVQYVDASRI VIKVNEDEMY PGEAGIDIYN LTKYTRSNQN TCINQMPCVS LGEPVERGDV LADGPSTDLG ELALGQNMR VAFMPWNGYN FEDSILVSER VVQEDRFTTI HIQELACVSR DTKLGPEEIT ADIPNVGEAA LSKLDESGIV YIGAEVTGGD ILVGKVTPK GETQLTPEEK LLRAIFGEKA SDVKDSSLRV PNGVSGTVID VQVFTRDGVE KDKRALEIEE MQLKQAKKDL S EELQILEA GLFSRIRAVL VAGGVEAEKL DKLPRDRWLE LGLTDEEKQN QLEQLAEQYD ELKHEFEKKL EAKRRKITQG DD LAPGVLK IVKVYLAVKR RIQPGDKMAG RHGNKGVISK INPIEDMPYD ENGTPVDIVL NPLGVPSRMN IGQILETHLG MAA KGIGDK INAMLKQQQE VAKLREFIQR AYDLGADVRQ KVDLSTFSDE EVMRLAENLR KGMPIATPVF DGAKEAEIKE LLKL GDLPT SGQIRLYDGR TGEQFERPVT VGYMYMLKLN HLVDDKMHAR STGSYSLVTQ QPLGGKAQFG GQRFGEMEVW ALEAY GAAY TLQEMLTVKS DDVNGRTKMY KNIVDGNHQM EPGMPESFNV LLKEIRSLGI NIELEDE UniProtKB: DNA-directed RNA polymerase subunit beta |
-Macromolecule #3: DNA-directed RNA polymerase subunit beta'
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta' / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 150.320219 KDa |
| Sequence | String: EEFDAIKIAL ASPDMIRSWS FGEVKKPETI NYRTFKPERD GLFCARIFGP VKDYECLCGK YKRLKHRGVI CEKCGVEVTQ TKVRRERMG HIELASPTAH IWFLKSLPSR IGLLLDMPLR DIERVLYFES YVVIEGGMTN LERQQILTEE QYLDALEEFG D EFDAKMGA ...String: EEFDAIKIAL ASPDMIRSWS FGEVKKPETI NYRTFKPERD GLFCARIFGP VKDYECLCGK YKRLKHRGVI CEKCGVEVTQ TKVRRERMG HIELASPTAH IWFLKSLPSR IGLLLDMPLR DIERVLYFES YVVIEGGMTN LERQQILTEE QYLDALEEFG D EFDAKMGA EAIQALLKSM DLEQECEQLR EELNETNSET KRKKLTKRIK LLEAFVQSGN KPEWMILTVL PVLPPDLRPL VP LDGGRFA TSDLNDLYRR VINRNNRLKR LLDLAAPDII VRNEKRMLQE AVDALLDNGR RGRAITGSNK RPLKSLADMI KGK QGRFRQ NLLGKRVDYS GRSVITVGPY LRLHQCGLPK KMALELFKPF IYGKLELRGL ATTIKAAKKM VEREEAVVWD ILDE VIREH PVLLNRAPTL HRLGIQAFEP VLIEGKAIQL HPLVCAAYNA DFDGDQMAVH VPLTLEAQLE ARALMMSTNN ILSPA NGEP IIVPSQDVVL GLYYMTRDCV NAKGMVLTGP AERLYRSGLA SLHARVKVRI TEYEKDANGE LVAKTSLKDT TVGRAI LWM IVPKGLPYSI VNQALGKKAI SKMLNTCYRI LGLKPTVIFA DQIMYTGFAY AARSGASVGI DDMVIPEKKH EIISEAE AE VAEIQEQFQS GLVTAGERYN KVIDIWAAAN DRVSKAMMDN LQTETVINRD GQEEKQVSFN SIYMMADSGA RGSAAQIR Q LAGMRGLMAK PDGSIIETPI TANFREGLNV LQYFISTHGA RKGLADTALK TANSGYLTRR LVDVAQDLVV TEDDCGTHE GIMMTPVIEG GDVKEPLRDR VLGRVTAEDV LKPGTADILV PRNTLLHEQW CDLLEENSVD AVKVRSVVSC DTDFGVCAHC YGRDLARGH IINKGEAIGV IAAQSIGEPG TQLTMRTFHI GGAASRAAAE SSIQVKNKGS IKLSNVKSVV NSSGKLVITS R NTELKLID EFGRTKESYK VPYGAVLAKG DGEQVAGGET VANWDPHTMP VITEVSGFVR FTDMIDGQTI TRQTDELTGL SS LVVLDSA ERTAGGKDLR PALKIVDAQG NDVLIPGTDM PAQYFLPGKA IVQLEDGVQI SSGDTLARIP QESGGTKDIT GGL PRVADL FEARRPKEPA ILAEISGIVS FGKETKGKRR LVITPVDGSD PYEEMIPKWR QLNVFEGERV ERGDVISDGP EAPH DILRL RGVHAVTRYI VNEVQDVYRL QGVKINDKHI EVIVRQMLRK ATIVNAGSSD FLEGEQVEYS RVKIANRELE ANGKV GATY SRDLLGITKA SLATESFISA ASFQETTRVL TEAAVAGKRD ELRGLKENVI VGRLIPAGTG YAYHQDRMRR RAAG UniProtKB: DNA-directed RNA polymerase subunit beta' |
-Macromolecule #4: DNA-directed RNA polymerase subunit alpha
| Macromolecule | Name: DNA-directed RNA polymerase subunit alpha / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.275273 KDa |
| Sequence | String: TEFLKPRLVD IEQVSSTHAK VTLEPLERGF GHTLGNALRR ILLSSMPGCA VTEVEIDGVL HEYSTKEGVQ EDILEILLNL KGLAVRVQG KDEVILTLNK SGIGPVTAAD ITHDGDVEIV KPQHVICHLT DENASISMRI KVQRGRGYVP ASTRIHSEED E RPIGRLLV ...String: TEFLKPRLVD IEQVSSTHAK VTLEPLERGF GHTLGNALRR ILLSSMPGCA VTEVEIDGVL HEYSTKEGVQ EDILEILLNL KGLAVRVQG KDEVILTLNK SGIGPVTAAD ITHDGDVEIV KPQHVICHLT DENASISMRI KVQRGRGYVP ASTRIHSEED E RPIGRLLV DACYSPVERI AYNVEAARVE QRTDLDKLVI EMETNGTIDP EEAIRRAATI LAEQLEAFVD LRDVRQPEVK EE KPEFDPI LLRPVDDLEL TVRSANCLKA EAIHYIGDLV QRTEVELLKT PNLGKKSLTE IKDVLASRGL SLGMRLENW UniProtKB: DNA-directed RNA polymerase subunit alpha |
-Macromolecule #5: DNA-directed RNA polymerase subunit alpha
| Macromolecule | Name: DNA-directed RNA polymerase subunit alpha / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.400266 KDa |
| Sequence | String: TEFLKPRLVD IEQVSSTHAK VTLEPLERGF GHTLGNALRR ILLSSMPGCA VTEVEIDGVL HEYSTKEGVQ EDILEILLNL KGLAVRVQG KDEVILTLNK SGIGPVTAAD ITHDGDVEIV KPQHVICHLT DENASISMRI KVQRGRGYVP ASTRIHSEED E RPIGRLLV ...String: TEFLKPRLVD IEQVSSTHAK VTLEPLERGF GHTLGNALRR ILLSSMPGCA VTEVEIDGVL HEYSTKEGVQ EDILEILLNL KGLAVRVQG KDEVILTLNK SGIGPVTAAD ITHDGDVEIV KPQHVICHLT DENASISMRI KVQRGRGYVP ASTRIHSEED E RPIGRLLV DACYSPVERI AYNVEAARVE QRTDLDKLVI EMETNGTIDP EEAIRRAATI LAEQLEAFVD LRDVRQPEVK EE KPEFDPI LLLPVDDLEL TVRSANCLKA EAIHYIGDLV QRTEVELLKT PNLGKKSLTE IKDVLASRGL SLG UniProtKB: DNA-directed RNA polymerase subunit alpha |
-Macromolecule #6: DNA-directed RNA polymerase subunit omega
| Macromolecule | Name: DNA-directed RNA polymerase subunit omega / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.118352 KDa |
| Sequence | String: ARVTVQDAVE KIGNRFDLVL VAARRARQMQ VGGKDPLVPE ENDKTTVIAL REIEEGLINN QILDVRERQE QQEQEAAELQ AVTAIAEGR R UniProtKB: DNA-directed RNA polymerase subunit omega |
-Macromolecule #7: DNA (94-MER)
| Macromolecule | Name: DNA (94-MER) / type: dna / ID: 7 / Details: NT strand DNA (94-MER) / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage lambda (virus) Enterobacteria phage lambda (virus) |
| Molecular weight | Theoretical: 29.094617 KDa |
| Sequence | String: (DA)(DA)(DT)(DC)(DT)(DA)(DT)(DC)(DA)(DC) (DC)(DG)(DC)(DA)(DA)(DG)(DG)(DG)(DA)(DT) (DA)(DA)(DA)(DT)(DA)(DT)(DC)(DT)(DA) (DA)(DC)(DA)(DC)(DC)(DG)(DT)(DG)(DC)(DG) (DT) (DG)(DT)(DT)(DG)(DA)(DC) ...String: (DA)(DA)(DT)(DC)(DT)(DA)(DT)(DC)(DA)(DC) (DC)(DG)(DC)(DA)(DA)(DG)(DG)(DG)(DA)(DT) (DA)(DA)(DA)(DT)(DA)(DT)(DC)(DT)(DA) (DA)(DC)(DA)(DC)(DC)(DG)(DT)(DG)(DC)(DG) (DT) (DG)(DT)(DT)(DG)(DA)(DC)(DT)(DA) (DT)(DT)(DT)(DT)(DA)(DC)(DC)(DT)(DC)(DT) (DG)(DG) (DC)(DG)(DG)(DT)(DG)(DA)(DT) (DA)(DA)(DT)(DG)(DG)(DT)(DT)(DG)(DC)(DA) (DT)(DG)(DT) (DA)(DC)(DT)(DA)(DA)(DG) (DG)(DA)(DG)(DG)(DT)(DT)(DG)(DT) |
-Macromolecule #8: DNA (94-MER)
| Macromolecule | Name: DNA (94-MER) / type: dna / ID: 8 / Details: T strand DNA (94-MER) / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage lambda (virus) Enterobacteria phage lambda (virus) |
| Molecular weight | Theoretical: 28.890529 KDa |
| Sequence | String: (DA)(DC)(DA)(DA)(DC)(DC)(DT)(DC)(DC)(DT) (DT)(DA)(DG)(DT)(DA)(DC)(DA)(DT)(DG)(DC) (DA)(DA)(DC)(DC)(DA)(DT)(DT)(DA)(DT) (DC)(DA)(DC)(DC)(DG)(DC)(DC)(DA)(DG)(DA) (DG) (DG)(DT)(DA)(DA)(DA)(DA) ...String: (DA)(DC)(DA)(DA)(DC)(DC)(DT)(DC)(DC)(DT) (DT)(DA)(DG)(DT)(DA)(DC)(DA)(DT)(DG)(DC) (DA)(DA)(DC)(DC)(DA)(DT)(DT)(DA)(DT) (DC)(DA)(DC)(DC)(DG)(DC)(DC)(DA)(DG)(DA) (DG) (DG)(DT)(DA)(DA)(DA)(DA)(DT)(DA) (DG)(DT)(DC)(DA)(DA)(DC)(DA)(DC)(DG)(DC) (DA)(DC) (DG)(DG)(DT)(DG)(DT)(DT)(DA) (DG)(DA)(DT)(DA)(DT)(DT)(DT)(DA)(DT)(DC) (DC)(DC)(DT) (DT)(DG)(DC)(DG)(DG)(DT) (DG)(DA)(DT)(DA)(DG)(DA)(DT)(DT) |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.9 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: Negatively stained EM specimens were prepared using a 2% uranyl acetate solution. |
| Grid | Support film - Material: CARBON / Support film - topology: CONTINUOUS |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2100 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Initial fiting was done using manual fitting in Chimera. Yasara software was used for energy minimizaton of the DNA and RNAP coordinates model. |
| Refinement | Protocol: OTHER |
| Output model |  PDB-6n4c: |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)