+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7pky | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Half-vault structure | |||||||||
 Components Components | Major vault protein | |||||||||
 Keywords Keywords | STRUCTURAL PROTEIN / Transport | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein activation cascade / ERBB signaling pathway / Neutrophil degranulation / negative regulation of epidermal growth factor receptor signaling pathway / protein phosphatase binding / cytoskeleton / cell population proliferation / ribonucleoprotein complex / protein kinase binding / perinuclear region of cytoplasm ...protein activation cascade / ERBB signaling pathway / Neutrophil degranulation / negative regulation of epidermal growth factor receptor signaling pathway / protein phosphatase binding / cytoskeleton / cell population proliferation / ribonucleoprotein complex / protein kinase binding / perinuclear region of cytoplasm / identical protein binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 7.9 Å | |||||||||
 Authors Authors | Guerra, P. / Gonzalez-Alamos, M. / Llauro, A. / Casanas, A. / Querol-Audi, J. / de Pablo, P. / Verdaguer, N. | |||||||||
| Funding support |  Spain, 2items Spain, 2items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Symmetry disruption commits vault particles to disassembly. Authors: Pablo Guerra / María González-Alamos / Aida Llauró / Arnau Casañas / Jordi Querol-Audí / Pedro J de Pablo / Núria Verdaguer /  Abstract: Vaults are ubiquitous ribonucleoprotein particles involved in a diversity of cellular processes, with promising applications as nanodevices for delivery of multiple cargos. The vault shell is ...Vaults are ubiquitous ribonucleoprotein particles involved in a diversity of cellular processes, with promising applications as nanodevices for delivery of multiple cargos. The vault shell is assembled by the symmetrical association of multiple copies of the major vault protein that, initially, generates half vaults. The pairwise, anti-parallel association of two half vaults produces whole vaults. Here, using a combination of vault recombinant reconstitution and structural techniques, we characterized the molecular determinants for the vault opening process. This process commences with a relaxation of the vault waist, causing the expansion of the inner cavity. Then, local disengagement of amino-terminal domains at the vault midsection seeds a conformational change that leads to the aperture, facilitating access to the inner cavity where cargo is hosted. These results inform a hitherto uncharacterized step of the vault cycle and will aid current engineering efforts leveraging vault for tailored cargo delivery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7pky.cif.gz 7pky.cif.gz | 5.1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7pky.ent.gz pdb7pky.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  7pky.json.gz 7pky.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pk/7pky https://data.pdbj.org/pub/pdb/validation_reports/pk/7pky ftp://data.pdbj.org/pub/pdb/validation_reports/pk/7pky ftp://data.pdbj.org/pub/pdb/validation_reports/pk/7pky | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  13482MC  7pkrC  7pkzC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 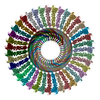
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
|
 Movie
Movie Controller
Controller





 PDBj
PDBj





 microscopy
microscopy