+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4867 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sub-tomogram averaging of Tula virus glycoprotein spike | |||||||||
 Map data Map data | Sub-tomogram averaging of Tula virus glycoprotein spike with corrected hand | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host TRAF-mediated signal transduction / host cell Golgi membrane / host cell mitochondrion / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / host cell surface / host cell endoplasmic reticulum membrane / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / fusion of virus membrane with host endosome membrane / viral envelope ...symbiont-mediated suppression of host TRAF-mediated signal transduction / host cell Golgi membrane / host cell mitochondrion / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MAVS activity / host cell surface / host cell endoplasmic reticulum membrane / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / virion membrane / signal transduction / zinc ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Tula orthohantavirus Tula orthohantavirus | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 15.6 Å | |||||||||
 Authors Authors | Huiskonen JT / Stass R / Li S | |||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: The Hantavirus Surface Glycoprotein Lattice and Its Fusion Control Mechanism. Authors: Alexandra Serris / Robert Stass / Eduardo A Bignon / Nicolás A Muena / Jean-Claude Manuguerra / Rohit K Jangra / Sai Li / Kartik Chandran / Nicole D Tischler / Juha T Huiskonen / Felix A ...Authors: Alexandra Serris / Robert Stass / Eduardo A Bignon / Nicolás A Muena / Jean-Claude Manuguerra / Rohit K Jangra / Sai Li / Kartik Chandran / Nicole D Tischler / Juha T Huiskonen / Felix A Rey / Pablo Guardado-Calvo /       Abstract: Hantaviruses are rodent-borne viruses causing serious zoonotic outbreaks worldwide for which no treatment is available. Hantavirus particles are pleomorphic and display a characteristic square ...Hantaviruses are rodent-borne viruses causing serious zoonotic outbreaks worldwide for which no treatment is available. Hantavirus particles are pleomorphic and display a characteristic square surface lattice. The envelope glycoproteins Gn and Gc form heterodimers that further assemble into tetrameric spikes, the lattice building blocks. The glycoproteins, which are the sole targets of neutralizing antibodies, drive virus entry via receptor-mediated endocytosis and endosomal membrane fusion. Here we describe the high-resolution X-ray structures of the heterodimer of Gc and the Gn head and of the homotetrameric Gn base. Docking them into an 11.4-Å-resolution cryoelectron tomography map of the hantavirus surface accounted for the complete extramembrane portion of the viral glycoprotein shell and allowed a detailed description of the surface organization of these pleomorphic virions. Our results, which further revealed a built-in mechanism controlling Gc membrane insertion for fusion, pave the way for immunogen design to protect against pathogenic hantaviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4867.map.gz emd_4867.map.gz | 14.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4867-v30.xml emd-4867-v30.xml emd-4867.xml emd-4867.xml | 7.8 KB 7.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4867.png emd_4867.png | 70.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4867 http://ftp.pdbj.org/pub/emdb/structures/EMD-4867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4867 | HTTPS FTP |
-Related structure data
| Related structure data |  6y5fC  6y5wC  6y62C  6y68C  6y6pC  6y6qC  6yrbC  6yrqC  6zjmC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4867.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4867.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sub-tomogram averaging of Tula virus glycoprotein spike with corrected hand | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
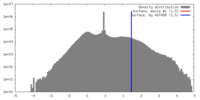
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tula orthohantavirus
| Entire | Name:  Tula orthohantavirus Tula orthohantavirus |
|---|---|
| Components |
|
-Supramolecule #1: Tula orthohantavirus
| Supramolecule | Name: Tula orthohantavirus / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 1980494 / Sci species name: Tula orthohantavirus / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: Yes / Virus empty: No |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 15.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: Dynamo / Number subtomograms used: 5449 |
|---|---|
| Extraction | Number tomograms: 30 / Number images used: 5449 |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)