[English] 日本語
 Yorodumi
Yorodumi- EMDB-21957: Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotaviru... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21957 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Reversed conformation | |||||||||
 Map data Map data | B-sharpened final map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / non-enveloped virus / viral particle / entry / membrane-penetration / rotavirus / VP4 / VP5* / VP8* / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral intermediate capsid / host cell endoplasmic reticulum lumen / host cell rough endoplasmic reticulum / permeabilization of host organelle membrane involved in viral entry into host cell / T=13 icosahedral viral capsid / host cytoskeleton / viral outer capsid / host cell endoplasmic reticulum-Golgi intermediate compartment / receptor-mediated virion attachment to host cell / host cell surface receptor binding ...viral intermediate capsid / host cell endoplasmic reticulum lumen / host cell rough endoplasmic reticulum / permeabilization of host organelle membrane involved in viral entry into host cell / T=13 icosahedral viral capsid / host cytoskeleton / viral outer capsid / host cell endoplasmic reticulum-Golgi intermediate compartment / receptor-mediated virion attachment to host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / structural molecule activity / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3]) Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3]) | |||||||||
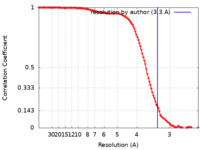
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Herrmann T / Harrison SC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Functional refolding of the penetration protein on a non-enveloped virus. Authors: Tobias Herrmann / Raúl Torres / Eric N Salgado / Cristina Berciu / Daniel Stoddard / Daniela Nicastro / Simon Jenni / Stephen C Harrison /  Abstract: A non-enveloped virus requires a membrane lesion to deliver its genome into a target cell. For rotaviruses, membrane perforation is a principal function of the viral outer-layer protein, VP4. Here we ...A non-enveloped virus requires a membrane lesion to deliver its genome into a target cell. For rotaviruses, membrane perforation is a principal function of the viral outer-layer protein, VP4. Here we describe the use of electron cryomicroscopy to determine how VP4 performs this function and show that when activated by cleavage to VP8* and VP5*, VP4 can rearrange on the virion surface from an 'upright' to a 'reversed' conformation. The reversed structure projects a previously buried 'foot' domain outwards into the membrane of the host cell to which the virion has attached. Electron cryotomograms of virus particles entering cells are consistent with this picture. Using a disulfide mutant of VP4, we have also stabilized a probable intermediate in the transition between the two conformations. Our results define molecular mechanisms for the first steps of the penetration of rotaviruses into the membranes of target cells and suggest similarities with mechanisms postulated for other viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21957.map.gz emd_21957.map.gz | 117 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21957-v30.xml emd-21957-v30.xml emd-21957.xml emd-21957.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21957_fsc.xml emd_21957_fsc.xml | 14.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_21957.png emd_21957.png | 101.2 KB | ||
| Masks |  emd_21957_msk_1.map emd_21957_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-21957.cif.gz emd-21957.cif.gz | 7.6 KB | ||
| Others |  emd_21957_half_map_1.map.gz emd_21957_half_map_1.map.gz emd_21957_half_map_2.map.gz emd_21957_half_map_2.map.gz | 98.3 MB 98.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21957 http://ftp.pdbj.org/pub/emdb/structures/EMD-21957 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21957 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21957 | HTTPS FTP |
-Related structure data
| Related structure data |  6wxgMC  6wxeC  6wxfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21957.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21957.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-sharpened final map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.231 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21957_msk_1.map emd_21957_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
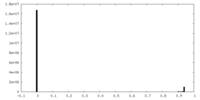
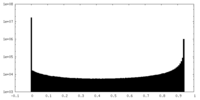
| Density Histograms |
-Half map: Half map1
| File | emd_21957_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
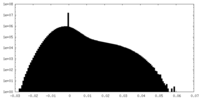
| Density Histograms |
-Half map: Half map2
| File | emd_21957_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
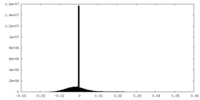
| Density Histograms |
- Sample components
Sample components
-Entire : Rotavirus VP4, VP6, VP7 assembly in the foldback conformation
| Entire | Name: Rotavirus VP4, VP6, VP7 assembly in the foldback conformation |
|---|---|
| Components |
|
-Supramolecule #1: Rotavirus VP4, VP6, VP7 assembly in the foldback conformation
| Supramolecule | Name: Rotavirus VP4, VP6, VP7 assembly in the foldback conformation type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: Obtained from wild-type recoated rhesus rotavirus particles (wt rcTLP) |
|---|---|
| Source (natural) | Organism:  Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3]) Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3]) |
| Molecular weight | Theoretical: 1.7 MDa |
-Macromolecule #1: Outer capsid protein VP4
| Macromolecule | Name: Outer capsid protein VP4 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3]) Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3])Strain: RVA/Monkey/United States/RRV/1975/G3P5B[3] |
| Molecular weight | Theoretical: 86.655586 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASLIYRQLL TNSYTVDLSD EIQEIGSTKT QNVTINLGPF AQTGYAPVNW GPGETNDSTT VEPVLDGPYQ PTTFNPPVDY WMLLAPTAA GVVVEGTNNT DRWLATILVE PNVTSETRSY TLFGTQEQIT IANASQTQWK FIDVVKTTQN GSYSQYGPLQ S TPKLYAVM ...String: MASLIYRQLL TNSYTVDLSD EIQEIGSTKT QNVTINLGPF AQTGYAPVNW GPGETNDSTT VEPVLDGPYQ PTTFNPPVDY WMLLAPTAA GVVVEGTNNT DRWLATILVE PNVTSETRSY TLFGTQEQIT IANASQTQWK FIDVVKTTQN GSYSQYGPLQ S TPKLYAVM KHNGKIYTYN GETPNVTTKY YSTTNYDSVN MTAFCDFYII PREEESTCTE YINNGLPPIQ NTRNIVPLAL SA RNIISHR AQANEDIVVS KTSLWKEMQY NRDITIRFKF ASSIVKSGGL GYKWSEISFK PANYQYTYTR DGEEVTAHTT CSV NGMNDF NFNGGSLPTD FVISRYEVIK ENSYVYVDYW DDSQAFRNMV YVRSLAANLN SVICTGGDYS FALPVGQWPV MTGG AVSLH SAGVTLSTQF TDFVSLNSLR FRFRLTVEEP SFSITRTRVS RLYGLPAANP NNGKEYYEVA GRFSLISLVP SNDDY QTPI TNSVTVRQDL ERQLGELREE FNALSQEIAM SQLIDLALLP LDMFSMFSGI KSTIDAAKSM ATSVMKKFKK SGLANS VST LTDSLSDAAS SISRGASIRS VGSSASAWTD VSTQITDVSS SVSSISTQTS TISRRLRLKE MATQTEGMNF DDISAAV LK TKIDRSTQIS PNTLPDIVTE ASEKFIPNRA YRVINNDEVF EAGTDGRFFA YRVETFDEIP FDVQKFADLV TDSPVISA I IDFKTLKNLN DNYGISRQQA FNLLRSDPRV LREFINQDNP IIRNRIEQLI MQCRL UniProtKB: Outer capsid protein VP4 |
-Macromolecule #2: Intermediate capsid protein VP6
| Macromolecule | Name: Intermediate capsid protein VP6 / type: protein_or_peptide / ID: 2 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3]) Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3])Strain: RVA/Monkey/United States/RRV/1975/G3P5B[3] |
| Molecular weight | Theoretical: 44.934766 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDVLYSLSKT LKDARDKIVE GTLYSNVSDL IQQFNQMIIT MNGNEFQTGG IGNLPIRNWN FDFGLLGTTL LNLDANYVET ARNTIDYFV DFVDNVCMDE MVRESQRNGI APQSDSLRKL SGIKFKRINF DNSSEYIENW NLQNRRQRTG FTFHKPNIFP Y SASFTLNR ...String: MDVLYSLSKT LKDARDKIVE GTLYSNVSDL IQQFNQMIIT MNGNEFQTGG IGNLPIRNWN FDFGLLGTTL LNLDANYVET ARNTIDYFV DFVDNVCMDE MVRESQRNGI APQSDSLRKL SGIKFKRINF DNSSEYIENW NLQNRRQRTG FTFHKPNIFP Y SASFTLNR SQPAHDNLMG TMWLNAGSEI QVAGFDYSCA INAPANIQQF EHIVQLRRVL TTATITLLPD AERFSFPRVI NS ADGATTW YFNPVILRPN NVEVEFLLNG QIINTYQARF GTIIARNFDT IRLSFQLMRP PNMTPAVAAL FPNAQPFEHH ATV GLTLRI ESAVCESVLA DASKTMLANV TSVRQEYAIP VGPVFPPGMN WTDLITNYSP SREDNLQRVF TVASIRSMLV K UniProtKB: Intermediate capsid protein VP6 |
-Macromolecule #3: Outer capsid glycoprotein VP7
| Macromolecule | Name: Outer capsid glycoprotein VP7 / type: protein_or_peptide / ID: 3 / Number of copies: 18 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3]) Rotavirus A (strain RVA/Monkey/United States/RRV/1975/G3P5B[3])Strain: RVA/Monkey/United States/RRV/1975/G3P5B[3] |
| Molecular weight | Theoretical: 37.136531 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYGIEYTTVL TFLISLILLN YILKSLTRMM DFIIYRFLFI VVILSPLLKA QNYGINLPIT GSMDTAYANS TQEETFLTST LCLYYPTEA ATEINDNSWK DTLSQLFLTK GWPTGSVYFK EYTDIASFSV DPQLYCDYNV VLMKYDATLQ LDMSELADLI L NEWLCNPM ...String: MYGIEYTTVL TFLISLILLN YILKSLTRMM DFIIYRFLFI VVILSPLLKA QNYGINLPIT GSMDTAYANS TQEETFLTST LCLYYPTEA ATEINDNSWK DTLSQLFLTK GWPTGSVYFK EYTDIASFSV DPQLYCDYNV VLMKYDATLQ LDMSELADLI L NEWLCNPM DITLYYYQQT DEANKWISMG SSCTIKVCPL NTQTLGIGCL TTDTATFEEV ATAEKLVITD VVDGVNHKLD VT TATCTIR NCKKLGPREN VAVIQVGGSD VLDITADPTT APQTERMMRI NWKKWWQVFY TVVDYVNQII QAMSKRSRSL NSA AFYYRI UniProtKB: Outer capsid glycoprotein VP7 |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 18 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 5 / Number of copies: 54 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: C-flat-1.2/1.3 4C / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 298.15 K / Instrument: GATAN CRYOPLUNGE 3 / Details: 4 ul sample volume, 4 sec blotting time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3701 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 1 / Number real images: 4107 / Average exposure time: 10.0 sec. / Average electron dose: 33.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 40605 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 27500 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | phenix.real_space_refine | ||||||
| Refinement | Protocol: OTHER | ||||||
| Output model |  PDB-6wxg: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)