4H8K
 
 | | Crystal structure of LC11-RNase H1 in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3'), Ribonuclease H | | Authors: | Nguyen, T.N, You, D.J, Matsumoto, H, Kanaya, E, Kanaya, S. | | Deposit date: | 2012-09-23 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of metagenome-derived LC11-RNase H1 in complex with RNA/DNA hybrid
J.Struct.Biol., 182, 2013
|
|
3U3G
 
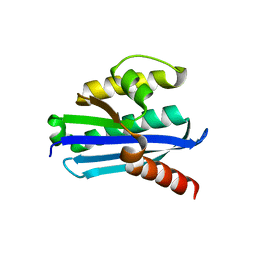 | | Structure of LC11-RNase H1 Isolated from Compost by Metagenomic Approach: Insight into the Structural Bases for Unusual Enzymatic Properties of Sto-RNase H1 | | Descriptor: | CHLORIDE ION, Ribonuclease H, UNKNOWN LIGAND | | Authors: | Nguyen, T.N, Angkawidjaja, C, Kanaya, E, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-10-05 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Activity, stability, and structure of metagenome-derived LC11-RNase H1, a homolog of Sulfolobus tokodaii RNase H1
Protein Sci., 21, 2012
|
|
5MTF
 
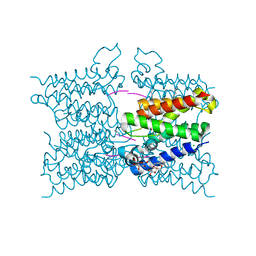 | | A modular route to novel potent and selective inhibitors of rhomboid intramembrane proteases | | Descriptor: | CHLORIDE ION, Rhomboid protease GlpG, inhibitor, ... | | Authors: | Ticha, A, Stanchev, S, Vinothkumar, K.R, Mikles, D.C, Pachl, P, Svehlova, K, Nguyen, M.T.N, Verhelst, S.H.L, Johnson, D, Bachovchin, D, Lepsik, M, Majer, P, Strisovsky, K. | | Deposit date: | 2017-01-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | General and Modular Strategy for Designing Potent, Selective, and Pharmacologically Compliant Inhibitors of Rhomboid Proteases.
Cell Chem Biol, 24, 2017
|
|
6D9H
 
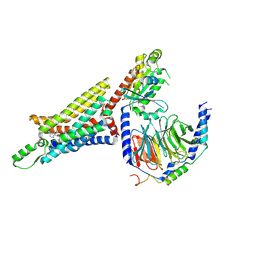 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Khoshouei, M, Thal, D.M, Liang, Y.-L, Nguyen, A.T.N, Furness, S.G.B, Venugopal, H, Baltos, J, Plitzko, J.M, Danev, R, Baumeister, W, May, L.T, Wootten, D, Sexton, P, Glukhova, A, Christopoulos, A. | | Deposit date: | 2018-04-29 | | Release date: | 2018-06-20 | | Last modified: | 2018-07-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the adenosine-bound human adenosine A1receptor-Gicomplex.
Nature, 558, 2018
|
|
3KRS
 
 | |
4Z55
 
 | |
8UYI
 
 | | Structure of ADP-bound and phosphorylated Pediculus humanus (Ph) PINK1 dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Serine/threonine-protein kinase Pink1, ... | | Authors: | Gan, Z.Y, Kirk, N.S, Leis, A, Komander, D. | | Deposit date: | 2023-11-13 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Interaction of PINK1 with nucleotides and kinetin.
Sci Adv, 10, 2024
|
|
8UYF
 
 | | Structure of nucleotide-free Pediculus humanus (Ph) PINK1 dimer | | Descriptor: | Serine/threonine-protein kinase Pink1, mitochondrial | | Authors: | Gan, Z.Y, Kirk, N.S, Leis, A, Komander, D. | | Deposit date: | 2023-11-13 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Interaction of PINK1 with nucleotides and kinetin.
Sci Adv, 10, 2024
|
|
8UYH
 
 | | Structure of AMP-PNP-bound Pediculus humanus (Ph) PINK1 dimer | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Pink1, ... | | Authors: | Gan, Z.Y, Kirk, N.S, Leis, A, Komander, D. | | Deposit date: | 2023-11-13 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Interaction of PINK1 with nucleotides and kinetin.
Sci Adv, 10, 2024
|
|
7JHX
 
 | | Crystal structure of hEPG5 LIR/GABARAPL1 complex | | Descriptor: | Ectopic P granules protein 5 homolog, Gamma-aminobutyric acid receptor-associated protein-like 1, SULFATE ION | | Authors: | Cheung, Y.W.S, Yip, C.K. | | Deposit date: | 2020-07-21 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Insights on autophagosome-lysosome tethering from structural and biochemical characterization of human autophagy factor EPG5.
Commun Biol, 4, 2021
|
|
8D28
 
 | |
8D2B
 
 | |
8D2A
 
 | |
8D5O
 
 | |
8D5L
 
 | | Crystal structure of theophylline aptamer in complex with TAL1 | | Descriptor: | 4-amino-8-methylpteridine-2,7(1H,8H)-dione, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Menichelli, E, Spraggon, G. | | Deposit date: | 2022-06-05 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of small molecules that target a tertiary-structured RNA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D29
 
 | |
8DO8
 
 | | Crystal structure ATG9 HDIR in complex with the ATG13:ATG101 HORMA dimer | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13, GLYCEROL | | Authors: | Buffalo, C.Z, Ren, X, Yokom, A.L, Hurley, J.H. | | Deposit date: | 2022-07-12 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for ATG9A recruitment to the ULK1 complex in mitophagy initiation.
Sci Adv, 9, 2023
|
|
8DK7
 
 | |
5MT7
 
 | |
5MT6
 
 | |
5MT8
 
 | |
6Y5S
 
 | | Crystal structure of savinase at cryogenic conditions | | Descriptor: | CALCIUM ION, SODIUM ION, Subtilisin Savinase | | Authors: | Wu, S, Moroz, O, Turkenburg, J, Nielsen, J.E, Wilson, K.S, Teilum, K. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Conformational heterogeneity of Savinase from NMR, HDX-MS and X-ray diffraction analysis.
Peerj, 8, 2020
|
|
5LK6
 
 | |
7LD3
 
 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist and an allosteric ligand | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Danev, R, Thal, D.M, Christopoulos, A, Glukhova, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-08 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Positive allosteric mechanisms of adenosine A 1 receptor-mediated analgesia.
Nature, 597, 2021
|
|
7LD4
 
 | | Cryo-EM structure of the human adenosine A1 receptor-Gi2-protein complex bound to its endogenous agonist | | Descriptor: | ADENOSINE, Chimera protein of Muscarinic acetylcholine receptor M4 and Adenosine receptor A1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Draper-Joyce, C.J, Danev, R, Thal, D.M, Christopoulos, A, Glukhova, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-09-08 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Positive allosteric mechanisms of adenosine A 1 receptor-mediated analgesia.
Nature, 597, 2021
|
|
