3DV2
 
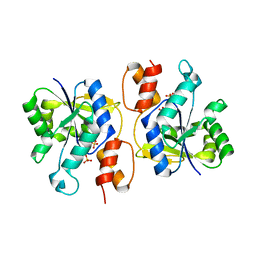 | | Crystal Structure of nicotinic acid mononucleotide adenylyltransferase from Bacillus anthracis | | Descriptor: | Nicotinate (Nicotinamide) nucleotide adenylyltransferase, SULFATE ION | | Authors: | Lu, S, Smith, C.D, Yang, Z, Pruett, P.S, Nagy, L, McCombs, D.P, DeLucas, L.J, Brouillette, W.J, Brouillette, C.G. | | Deposit date: | 2008-07-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of nicotinic acid mononucleotide adenylyltransferase from Bacillus anthracis.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
1XHL
 
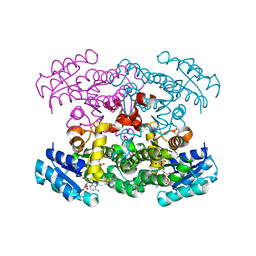 | | Crystal Structure of putative Tropinone Reductase-II from Caenorhabditis Elegans with Cofactor and Substrate | | Descriptor: | 8-METHYL-8-AZABICYCLO[3,2,1]OCTAN-3-ONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase family member (5L265), ... | | Authors: | Schormann, N, Karpova, E, Zhou, J, Zhang, Y, Symersky, J, Bunzel, R, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKInstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of putative Tropinone Reductase-II from Caenorhabditis Elegans with Cofactor and Substrate
To be Published
|
|
1XKQ
 
 | | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, short-chain reductase family member (5D234) | | Authors: | Schormann, N, Zhou, J, Karpova, E, Zhang, Y, Symersky, J, Bunzel, B, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKinstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-29 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor
To be Published
|
|
1SCZ
 
 | | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase | | Descriptor: | Dihydrolipoamide Succinyltransferase | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Tsao, J, Johnson, D, Huang, W.-Y, Pruett, P, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, Z, Lu, S, DeLucas, L. | | Deposit date: | 2004-02-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved structural model for the catalytic domain of E.coli dihydrolipoamide succinyltransferase
To be Published
|
|
1SPX
 
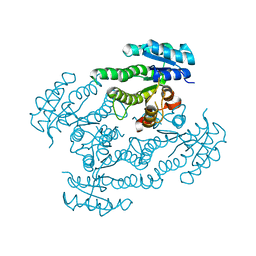 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | Descriptor: | short-chain reductase family member (5L265) | | Authors: | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
2XTD
 
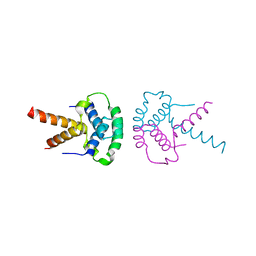 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | TBL1 F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1H9U
 
 | | The structure of the human retinoid-X-receptor beta ligand binding domain in complex with the specific synthetic agonist LG100268 | | Descriptor: | 6-[1-(3,5,5,8,8-PENTAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)CYCLOPROPYL]PYRIDINE-3-CARBOXYLIC ACID, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Schwabe, J.W.R, Love, J.D, Gooch, J.T. | | Deposit date: | 2001-03-21 | | Release date: | 2002-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structural Basis for the Specificity of Retinoid-X Receptor-Selective Agonists: New Insights Into the Role of Helix H12
J.Biol.Chem., 277, 2002
|
|
2XTC
 
 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2XTE
 
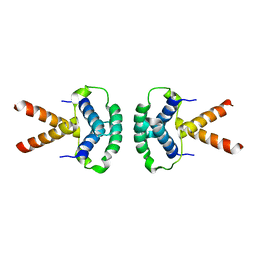 | | Structure of the TBL1 tetramerisation domain | | Descriptor: | F-BOX-LIKE/WD REPEAT-CONTAINING PROTEIN TBL1X | | Authors: | Oberoi, J, Fairall, L, Watson, P.J, Greenwood, J.A, Schwabe, J.W.R. | | Deposit date: | 2010-10-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis for the Assembly of the Smrt/Ncor Core Transcriptional Repression Machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8TQE
 
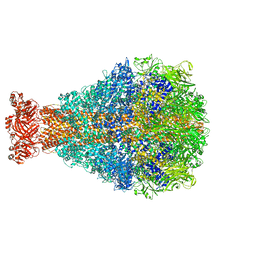 | | XptA2 wild type | | Descriptor: | XptA2 | | Authors: | Martin, C.L, Binshtein, E.M, Aller, S.G. | | Deposit date: | 2023-08-07 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the Insecticidal Toxin Complex Subunit XptA2 Highlight Roles for Flexible Domains.
Int J Mol Sci, 24, 2023
|
|
8TV0
 
 | | XptA2 wild type | | Descriptor: | XptA2 | | Authors: | Martin, C.L, Aller, S.G. | | Deposit date: | 2023-08-17 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the Insecticidal Toxin Complex Subunit XptA2 Highlight Roles for Flexible Domains.
Int J Mol Sci, 24, 2023
|
|
2L5G
 
 | | Co-ordinates and 1H, 13C and 15N chemical shift assignments for the complex of GPS2 53-90 and SMRT 167-207 | | Descriptor: | G protein pathway suppressor 2, Putative uncharacterized protein NCOR2 | | Authors: | Oberoi, J, Yang, J, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2010-11-01 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4ZSH
 
 | | RXR LBD in complex with 9-cis-13,14-dihydroretinoic acid | | Descriptor: | (5S,6S,9R,13R)-2,3-didehydro-5,6,7,8,9,10,11,12,13,14-decahydroretinoic acid, NCoA2 peptide, Retinoic acid receptor RXR-alpha | | Authors: | Rochel, N, Krezel, W, Ruhl, R. | | Deposit date: | 2015-05-13 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 9-cis-13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice.
Plos Genet., 11, 2015
|
|
1PZV
 
 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Probable ubiquitin-conjugating enzyme E2-19 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-14 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
2VSR
 
 | | hPPARgamma Ligand binding domain in complex with 9-(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV1
 
 | | hPPARgamma Ligand binding domain in complex with 4-HDHA | | Descriptor: | (4S,5E,7Z,10Z,13Z,16Z,19Z)-4-hydroxydocosa-5,7,10,13,16,19-hexaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV2
 
 | | hPPARgamma Ligand binding domain in complex with 5-HEPA | | Descriptor: | (5R,6E,8Z,11Z,14Z,17Z)-5-hydroxyicosa-6,8,11,14,17-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV4
 
 | | hPPARgamma Ligand binding domain in complex with 6-oxoOTE | | Descriptor: | (8E,10S,12Z)-10-hydroxy-6-oxooctadeca-8,12-dienoic acid, (8R,9Z,12Z)-8-hydroxy-6-oxooctadeca-9,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VST
 
 | | hPPARgamma Ligand binding domain in complex with 13-(S)-HODE | | Descriptor: | (9Z,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV3
 
 | | hPPARgamma Ligand binding domain in complex with 4-oxoDHA | | Descriptor: | (6E,10Z,13Z,16Z,19Z)-4-oxodocosa-6,10,13,16,19-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation of Ppargamma by Oxidized Fatty Acids.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV0
 
 | | hPPARgamma Ligand binding domain in complex with DHA | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
1R9H
 
 | |
1ROW
 
 | | Structure of SSP-19, an MSP-domain protein like family member in Caenorhabditis elegans | | Descriptor: | MSP-domain protein like family member | | Authors: | Schormann, N, Symersky, J, Carson, M, Luo, M, Lin, G, Li, S, Qiu, S, Arabashi, A, Bunzel, B, Luo, D, Nagy, L, Gray, R, Luan, C.-H, Zhang, J, Lu, S, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-02 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of sperm-specific protein SSP-19 from Caenorhabditis elegans.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OOE
 
 | | Structural Genomics of Caenorhabditis elegans : Dihydropteridine reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydropteridine reductase | | Authors: | Symersky, J, Li, S, Nagy, L, Qiu, S, Lin, G, Tsao, J, Luo, D, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of dihydropteridine reductase.
Proteins, 53, 2003
|
|
