1N3Y
 
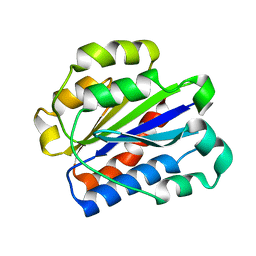 | | Crystal structure of the alpha-X beta2 integrin I domain | | Descriptor: | Integrin alpha-X | | Authors: | Vorup-Jensen, T, Ostermeier, C, Shimaoka, M, Hommel, U, Springer, T.A. | | Deposit date: | 2002-10-30 | | Release date: | 2003-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and allosteric regulation of the alpha X beta 2 integrin I domain.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1CQP
 
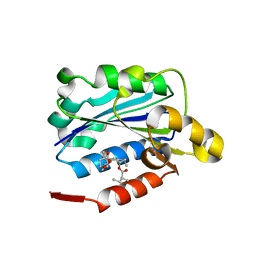 | | CRYSTAL STRUCTURE ANALYSIS OF THE COMPLEX LFA-1 (CD11A) I-DOMAIN / LOVASTATIN AT 2.6 A RESOLUTION | | Descriptor: | ANTIGEN CD11A (P180), LOVASTATIN, MAGNESIUM ION | | Authors: | Kallen, J, Welzenbach, K, Ramage, P, Geyl, D, Kriwacki, R, Legge, G, Cottens, S, Weitz-Schmidt, G, Hommel, U. | | Deposit date: | 1999-08-10 | | Release date: | 2000-08-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for LFA-1 inhibition upon lovastatin binding to the CD11a I-domain.
J.Mol.Biol., 292, 1999
|
|
1DGQ
 
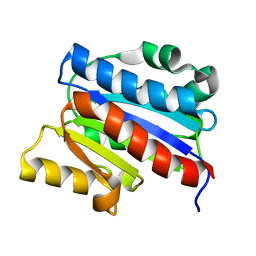 | | NMR SOLUTION STRUCTURE OF THE INSERTED DOMAIN OF HUMAN LEUKOCYTE FUNCTION ASSOCIATED ANTIGEN-1 | | Descriptor: | LEUKOCYTE FUNCTION ASSOCIATED ANTIGEN-1 | | Authors: | Legge, G.B, Kriwacki, R.W, Chung, J, Hommel, U, Ramage, P, Case, D.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 1999-11-24 | | Release date: | 2000-02-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the inserted domain of human leukocyte function associated antigen-1.
J.Mol.Biol., 295, 2000
|
|
1XUO
 
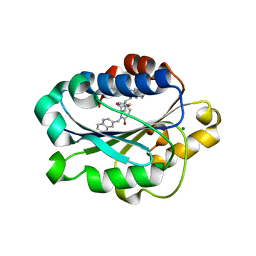 | | X-ray structure of LFA-1 I-domain bound to a 1,4-diazepane-2,5-dione inhibitor at 1.8A resolution | | Descriptor: | (2R)-2-[3-ISOBUTYL-2,5-DIOXO-4-(QUINOLIN-3-YLMETHYL)-1,4-DIAZEPAN-1-YL]-N-METHYL-3-(2-NAPHTHYL)PROPANAMIDE, Integrin alpha-L, MAGNESIUM ION | | Authors: | Wattanasin, S, Kallen, J, Myers, S, Guo, Q, Sabio, M, Ehrhardt, C, Albert, R, Hommel, U, Weckbecker, G, Welzenbach, K. | | Deposit date: | 2004-10-26 | | Release date: | 2005-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,4-Diazepane-2,5-diones as novel inhibitors of LFA-1
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8C3U
 
 | | Crystal Structure of human IL-1beta in complex with a low molecular weight antagonist | | Descriptor: | (S)-4'-hydroxy-3'-(6-methyl-2-oxo-3-(1H-pyrazol-4-yl)indolin-3-yl)-[1,1'-biphenyl]-2,4-dicarboxylic acid, Interleukin-1 beta | | Authors: | Rondeau, J.-M, Lehmann, S, Koch, E. | | Deposit date: | 2022-12-28 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Discovery of a selective and biologically active low-molecular weight antagonist of human interleukin-1 beta.
Nat Commun, 14, 2023
|
|
8RYS
 
 | | Human IL-1beta, unliganded | | Descriptor: | Interleukin-1 beta, SULFATE ION | | Authors: | Rondeau, J.-M, Lehmann, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Ligandability assessment of IL-1beta by integrated hit identification approaches
To be published
|
|
2EWY
 
 | |
8F1G
 
 | | Crystal structure of human WDR5 in complex with compound WM662 | | Descriptor: | (2S)-2-({(2S)-3-(3'-chloro[1,1'-biphenyl]-4-yl)-1-oxo-1-[(1H-tetrazol-5-yl)amino]propan-2-yl}oxy)propanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Liu, H. | | Deposit date: | 2022-11-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of WDR5-MYC Interaction.
Acs Chem.Biol., 18, 2023
|
|
2FP7
 
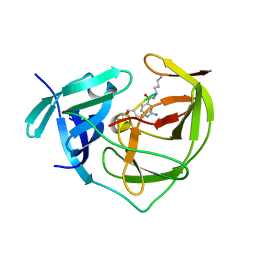 | |
2FOM
 
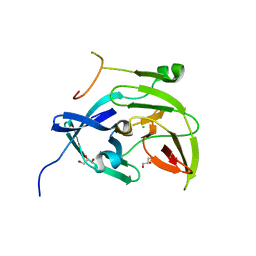 | | Dengue Virus NS2B/NS3 Protease | | Descriptor: | CHLORIDE ION, GLYCEROL, polyprotein | | Authors: | Schiering, N, Kroemer, M, Renatus, M, Erbel, P, D'Arcy, A. | | Deposit date: | 2006-01-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1MIL
 
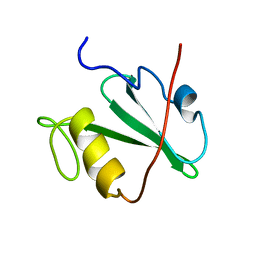 | | TRANSFORMING PROTEIN | | Descriptor: | SHC ADAPTOR PROTEIN | | Authors: | Mikol, V. | | Deposit date: | 1995-09-20 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the SH2 domain from the adaptor protein SHC: a model for peptide binding based on X-ray and NMR data.
J.Mol.Biol., 254, 1995
|
|
5FCR
 
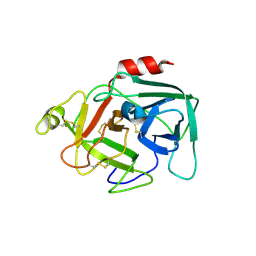 | | MOUSE COMPLEMENT FACTOR D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Complement factor D, DIMETHYL SULFOXIDE, ... | | Authors: | Mac Sweeney, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FBE
 
 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND2 | | Descriptor: | Complement factor D, GLYCEROL, methyl 2-[[[(2~{S})-2-[[3-(trifluoromethyloxy)phenyl]carbamoyl]pyrrolidin-1-yl]carbonylamino]methyl]benzoate | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FAH
 
 | | KALLIKREIN-7 IN COMPLEX WITH COMPOUND1 | | Descriptor: | (2~{S})-~{N}2-[2-(4-methoxyphenyl)ethyl]-~{N}1-(naphthalen-1-ylmethyl)pyrrolidine-1,2-dicarboxamide, ACETATE ION, Kallikrein-7 | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FCK
 
 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[[(1~{R})-1-(3-chloranyl-2-fluoranyl-phenyl)ethyl]carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]pyrazolo[3,4-c]pyridine-3-carboxamide, Complement factor D, SULFATE ION | | Authors: | Mac Sweeney, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5FBI
 
 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND 3b | | Descriptor: | 3-[(2-aminocarbonyl-1~{H}-indol-5-yl)oxymethyl]benzoic acid, Complement factor D, GLYCEROL | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
5MVS
 
 | | Crystal structure of Dot1L in complex with adenosine and inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] | | Descriptor: | ADENOSINE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-17 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MW4
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD7 [N-(3-(((R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl)(methyl)amino)propyl)-2-(3-(2-chloro-3-(2-methylpyridin-3-yl)benzo[b]thiophen-5-yl)ureido)acetamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~2~-{[2-chloro-3-(2-methylpyridin-3-yl)-1-benzothiophen-5-yl]carbamoyl}-N-(3-{methyl[(3R)-1-(5H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]amino}propyl)glycinamide, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MT0
 
 | |
5MW3
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] and inhibitor CPD2 [(R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine] | | Descriptor: | (3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MT4
 
 | |
5NB7
 
 | | Complement factor D | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[(6-bromanylpyridin-2-yl)carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]indazole-3-carboxamide, Complement factor D, DIMETHYL SULFOXIDE | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NAR
 
 | |
5NAW
 
 | | Complement factor D in complex with the inhibitor (1R,3S,5R)-2-Aza-bicyclo[3.1.0]hexane-2,3-dicarboxylic acid 2-[(1-carbamoyl-1H-indol-3-yl)-amide] 3-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (1~{R},3~{S},5~{R})-~{N}2-(1-aminocarbonylindol-3-yl)-~{N}3-[3-(trifluoromethyloxy)phenyl]-2-azabicyclo[3.1.0]hexane-2,3-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-02-28 | | Release date: | 2017-06-28 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NBA
 
 | | Complement factor D in complex with the inhibitor (2S,4R)-4-Fluoro-pyrrolidine-1,2-dicarboxylic acid 1-[(1-carbamoyl-1H-indol-3-yl)-amide] 2-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (2~{S},4~{R})-~{N}1-(1-aminocarbonylindol-3-yl)-4-fluoranyl-~{N}2-[3-(trifluoromethyloxy)phenyl]pyrrolidine-1,2-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
