2DRP
 
 | | THE CRYSTAL STRUCTURE OF A TWO ZINC-FINGER PEPTIDE REVEALS AN EXTENSION TO THE RULES FOR ZINC-FINGER/DNA RECOGNITION | | Descriptor: | DNA (5'-D(*CP*TP*AP*AP*TP*AP*AP*GP*GP*AP*TP*AP*AP*CP*GP*TP*C P*CP*G)-3'), DNA (5'-D(*TP*CP*GP*GP*AP*CP*GP*TP*TP*AP*TP*CP*CP*TP*TP*AP*T P*TP*A)-3'), PROTEIN (TRAMTRACK DNA-BINDING DOMAIN), ... | | Authors: | Fairall, L, Schwabe, J.W.R, Chapman, L, Finch, J.T, Rhodes, D. | | Deposit date: | 1994-06-06 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition.
Nature, 366, 1993
|
|
1H6O
 
 | | Dimerisation domain from human TRF1 | | Descriptor: | TELOMERIC REPEAT BINDING FACTOR 1 | | Authors: | Fairall, L, Chapman, L, Rhodes, D. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-05 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2.
Mol.Cell, 8, 2001
|
|
6Z2J
 
 | | The structure of the dimeric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
6Z2K
 
 | | The structure of the tetrameric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
2YHO
 
 | | The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor | | Descriptor: | ACETATE ION, E3 UBIQUITIN-PROTEIN LIGASE MYLIP, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Fairall, L, Goult, B.T, Millard, C.J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Idol-Ube2D Complex Mediates Sterol-Dependent Degradation of the Ldl Receptor.
Genes Dev., 25, 2011
|
|
1H6P
 
 | | Dimeristion domain from human TRF2 | | Descriptor: | MAGNESIUM ION, TELOMERIC REPEAT BINDING FACTOR 2 | | Authors: | Chapman, L, Fairall, L, Rhodes, D. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Trfh Dimerization Domain of the Human Telomere Proteins Trf1 and Trf2
Mol.Cell, 8, 2001
|
|
8AXW
 
 | | The structure of mouse AsterC (GramD1c) with Ezetimibe | | Descriptor: | (3~{R},4~{S})-1-(4-fluorophenyl)-3-[(3~{S})-3-(4-fluorophenyl)-3-oxidanyl-propyl]-4-(4-hydroxyphenyl)azetidin-2-one, CHLORIDE ION, ETHANOL, ... | | Authors: | Fairall, L, Xiao, X, Burger, L, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aster-dependent nonvesicular transport facilitates dietary cholesterol uptake.
Science, 382, 2023
|
|
6GQF
 
 | | The structure of mouse AsterA (GramD1a) with 25-hydroxy cholesterol | | Descriptor: | 25-HYDROXYCHOLESTEROL, GLYCEROL, GRAM domain-containing protein 1A | | Authors: | Fairall, L, Gurnett, J.E, Vashi, D, Sandhu, J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Aster Proteins Facilitate Nonvesicular Plasma Membrane to ER Cholesterol Transport in Mammalian Cells.
Cell, 175, 2018
|
|
2YHN
 
 | | The IDOL-UBE2D complex mediates sterol-dependent degradation of the LDL receptor | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE MYLIP, ZINC ION | | Authors: | Fairall, L, Goult, B.T, Millard, C.J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Idol-Ube2D Complex Mediates Sterol-Dependent Degradation of the Ldl Receptor.
Genes Dev., 25, 2011
|
|
4CP3
 
 | | The structure of BCL6 BTB (POZ) domain in complex with the ansamycin antibiotic rifabutin. | | Descriptor: | B-CELL LYMPHOMA 6 PROTEIN, RIFABUTIN | | Authors: | Evans, S.E, Fairall, L, Goult, B.T, Jamieson, A.G, Ferrigno, P.K, Ford, R, Wagner, S.D, Schwabe, J.W.R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Ansamycin Antibiotic, Rifamycin Sv, Inhibits Bcl6 Transcriptional Repression and Forms a Complex with the Bcl6-Btb/Poz Domain.
Plos One, 9, 2014
|
|
6G16
 
 | | Structure of the human RBBP4:MTA1(464-546) complex showing loop exchange | | Descriptor: | Histone-binding protein RBBP4, Metastasis-associated protein MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
7QDT
 
 | | Crystal structure of a mutant (P393GX) Thyroid Receptor Alpha ligand binding domain designed to model dominant negative human mutations. | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Isoform Alpha-1 of Thyroid hormone receptor alpha | | Authors: | Romartinez-Alonso, B, Fairall, L, Agostini, M, Chatterjee, K, Schwabe, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Guided Approach to Relieving Transcriptional Repression in Resistance to Thyroid Hormone alpha.
Mol.Cell.Biol., 42, 2022
|
|
5FXY
 
 | | Structure of the human RBBP4:MTA1(464-546) complex | | Descriptor: | HISTONE-BINDING PROTEIN RBBP4, METASTASIS-ASSOCIATED PROTEIN MTA1 | | Authors: | Millard, C.J, Varma, N, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Elife, 5, 2016
|
|
8AU4
 
 | | Structural insights reveal a heterotetramer between oncogenic K-Ras4BG12V and Rgl2, a RalA/B activator | | Descriptor: | Ral guanine nucleotide dissociation stimulator-like 2 | | Authors: | Tariq, M, Ikeya, T, Togashi, N, Fairall, L, Alejo, C.B, Kamei, S, Alonso, B.R, Campillo, M.A.M, Hudson, A, Ito, Y, Schwabe, J, Dominguez, C, Tanaka, K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
8B69
 
 | | Heterotetramer of K-Ras4B(G12V) and Rgl2(RBD) | | Descriptor: | Isoform 2B of GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Tariq, M, Fairall, L, Romartinez-Alonso, B, Dominguez, C, Schwabe, J.W.R, Tanaka, K. | | Deposit date: | 2022-09-26 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
1W0U
 
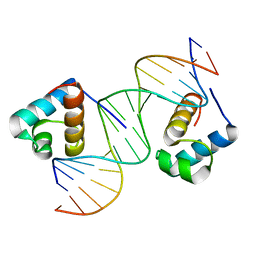 | | hTRF2 DNA-binding domain in complex with telomeric DNA. | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP *CP*CP*CP*TP*AP*GP*A)-3', 5'-D(*TP*CP*TP*AP*GP*GP*GP*TP*TP*AP *GP*GP*GP*TP*TP*AP*G)-3', TELOMERIC REPEAT BINDING FACTOR 2 | | Authors: | Court, R.I, Chapman, L.M, Fairall, L, Rhodes, D. | | Deposit date: | 2004-06-11 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How the Human Telomeric Proteins Trf1 and Trf2 Recognize Telomeric DNA: A View from High-Resolution Crystal Structures
Embo Rep., 6, 2005
|
|
1W0T
 
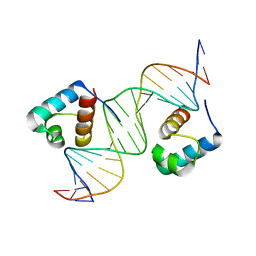 | | hTRF1 DNA-binding domain in complex with telomeric DNA. | | Descriptor: | 5'-D(*CP*TP*GP*TP*TP*AP*GP*GP*GP*TP *TP*AP*GP*GP*GP*TP*TP*AP*G)-3', 5'-D(*TP*CP*TP*AP*AP*CP*CP*CP*TP*AP *AP*CP*CP*CP*TP*AP*AP*CP*A)-3', TELOMERIC REPEAT BINDING FACTOR 1 | | Authors: | Court, R.I, Chapman, L.M, Fairall, L, Rhodes, D. | | Deposit date: | 2004-06-11 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How the Human Telomeric Proteins Trf1 and Trf2 Recognize Telomeric DNA: A View from High-Resolution Crystal Structures
Embo Rep., 6, 2005
|
|
4BKX
 
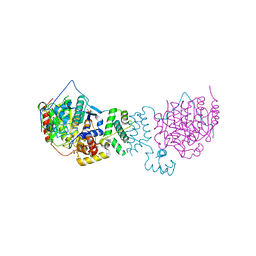 | | The structure of HDAC1 in complex with the dimeric ELM2-SANT domain of MTA1 from the NuRD complex | | Descriptor: | ACETATE ION, HISTONE DEACETYLASE 1, METASTASIS-ASSOCIATED PROTEIN MTA1, ... | | Authors: | Millard, C.J, Watson, P.J, Celardo, I, Gordiyenko, Y, Cowley, S.M, Robinson, C.V, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2013-04-30 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Class I Hdacs Share a Common Mechanism of Regulation by Inositol Phosphates.
Mol.Cell, 51, 2013
|
|
4A69
 
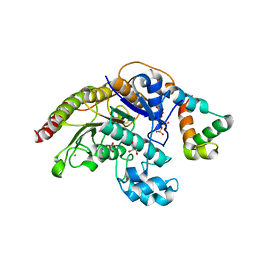 | | Structure of HDAC3 bound to corepressor and inositol tetraphosphate | | Descriptor: | ACETATE ION, D-MYO INOSITOL 1,4,5,6 TETRAKISPHOSPHATE, GLYCEROL, ... | | Authors: | Watson, P.J, Fairall, L, Santos, G.M, Schwabe, J.W.R. | | Deposit date: | 2011-11-01 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of Hdac3 Bound to Co-Repressor and Inositol Tetraphosphate.
Nature, 481, 2012
|
|
4D6K
 
 | | Structure of DNTTIP1 dimerisation domain. | | Descriptor: | DEOXYNUCLEOTIDYLTRANSFERASE TERMINAL-INTERACTING PROTEIN 1 | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of a Cell Cycle Associated Hdac1/2 Complex Reveals the Structural Basis for Complex Assembly and Nucleosome Targeting.
Nucleic Acids Res., 43, 2015
|
|
7AOA
 
 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AHJ
 
 | | Crystal structure of PPARgamma V290M mutant ligand binding domain in complex with farglitazar | | Descriptor: | 2-(2-BENZOYL-PHENYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Schoenmakers, E, Schwabe, B.T.W, Fairall, L, Chatterjee, K, Schwabe, J.W.R. | | Deposit date: | 2020-09-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PPARgamma V290M mutant ligand binding domain in complex with farglitazar
To Be Published
|
|
7AO9
 
 | | Structure of the core MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO8
 
 | | Structure of the MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
2VV1
 
 | | hPPARgamma Ligand binding domain in complex with 4-HDHA | | Descriptor: | (4S,5E,7Z,10Z,13Z,16Z,19Z)-4-hydroxydocosa-5,7,10,13,16,19-hexaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
