3Q48
 
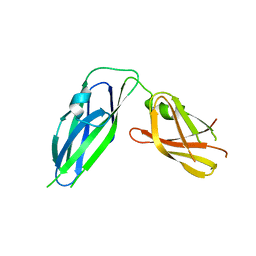 | | Crystal structure of Pseudomonas aeruginosa CupB2 chaperone | | Descriptor: | Chaperone CupB2 | | Authors: | Cai, X, Wang, R, Filloux, A, Waksman, G, Meng, G. | | Deposit date: | 2010-12-23 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of Pseudomonas aeruginosa CupB chaperones
Plos One, 6, 2011
|
|
4HRV
 
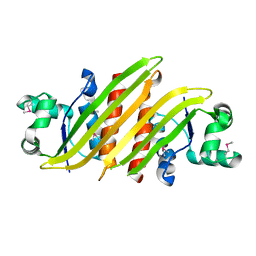 | |
7CU8
 
 | | Crystal structure of the soluble domain of TiME protein from Mycobacterium tuberculosis | | Descriptor: | SULFATE ION, Tube-forming protein in Mycobacterial Envelope (TiME) | | Authors: | Gong, W, Cai, X, Liu, L, Wen, C. | | Deposit date: | 2020-08-21 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Identification and architecture of a putative secretion tube across mycobacterial outer envelope.
Sci Adv, 7, 2021
|
|
7CU9
 
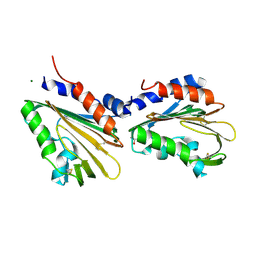 | | Crystal structure of the soluble domain of TiME protein from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, MAGNESIUM ION, Tube-forming protein in Mycobacterial Envelpe, ... | | Authors: | Gong, W, Cai, X, Liu, L, Wen, C. | | Deposit date: | 2020-08-21 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and architecture of a putative secretion tube across mycobacterial outer envelope.
Sci Adv, 7, 2021
|
|
1QAC
 
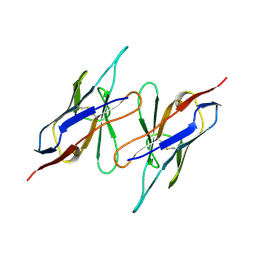 | | CHANGE IN DIMERIZATION MODE BY REMOVAL OF A SINGLE UNSATISFIED POLAR RESIDUE | | Descriptor: | IMMUNOGLOBULIN LIGHT CHAIN VARIABLE DOMAIN | | Authors: | Pokkuluri, P.R, Cai, X, Johnson, G, Stevens, F.J, Schiffer, M. | | Deposit date: | 1999-02-25 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Change in dimerization mode by removal of a single unsatisfied polar residue located at the interface.
Protein Sci., 9, 2000
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
6EWS
 
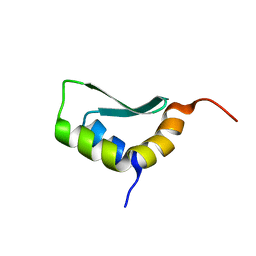 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12A-NDD | | Descriptor: | NRPS Kj12A-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWV
 
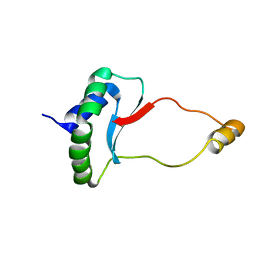 | | Solution Structure of Docking Domain Complex of RXP NRPS: Kj12C NDD - Kj12B CDD | | Descriptor: | NRPS Kj12C-NDD, NRPS Kj12B-CDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6IMQ
 
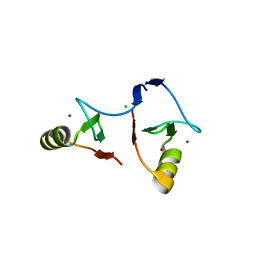 | | Crystal structure of PML B1-box multimers | | Descriptor: | CHLORIDE ION, Protein PML, ZINC ION | | Authors: | Li, Y, Ma, X, Chen, Z, Wu, H, Wang, P, Wu, W, Cheng, N, Zeng, L, Zhang, H, Cai, X, Chen, S.J, Chen, Z, Meng, G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | B1 oligomerization regulates PML nuclear body biogenesis and leukemogenesis.
Nat Commun, 10, 2019
|
|
1EEU
 
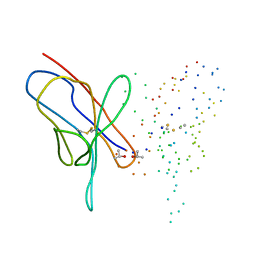 | | M4L/Y(27D)D/Q89D/T94H mutant of LEN | | Descriptor: | ISOPROPYL ALCOHOL, KAPPA-4 IMMUNOGLOBULIN (LIGHT CHAIN) | | Authors: | Pokkuluri, P.R, Cai, X, Gu, M, Stevens, F.J, Schiffer, M. | | Deposit date: | 2000-02-03 | | Release date: | 2001-02-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Factors contributing to decreased protein stability when aspartic acid residues are in beta-sheet regions.
Protein Sci., 11, 2002
|
|
1EFQ
 
 | | Q38D mutant of LEN | | Descriptor: | KAPPA-4 IMMUNOGLOBULIN (LIGHT CHAIN), URANYL (VI) ION, ZINC ION | | Authors: | Pokkuluri, P.R, Cai, X, Gu, M, Stevens, F.J, Schiffer, M. | | Deposit date: | 2000-02-09 | | Release date: | 2001-02-09 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Factors contributing to decreased protein stability when aspartic acid residues are in beta-sheet regions.
Protein Sci., 11, 2002
|
|
6EWT
 
 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12B-NDD | | Descriptor: | NRPS Kj12B-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6EWU
 
 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12C-NDD | | Descriptor: | NRPS Kj12C-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
3TEQ
 
 | | Crystal structure of SOAR domain | | Descriptor: | PHOSPHATE ION, Stromal interaction molecule 1 | | Authors: | Yang, X, Jin, H, Cai, X, Shen, Y. | | Deposit date: | 2011-08-15 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the activation of Stromal interaction molecule 1 (STIM1).
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3TER
 
 | |
5VEX
 
 | | Structure of the human GLP-1 receptor complex with NNC0640 | | Descriptor: | 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, Glucagon-like peptide 1 receptor, Endolysin chimera | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
5VEW
 
 | | Structure of the human GLP-1 receptor complex with PF-06372222 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin chimera, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine, ... | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
5V54
 
 | | Crystal structure of 5-HT1B receptor in complex with methiothepin | | Descriptor: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 1B,OB-1 fused 5-HT1b receptor,5-hydroxytryptamine receptor 1B | | Authors: | Yin, W.C, Zhou, X.E, Yang, D, de Waal, P, Wang, M.T, Dai, A, Cai, X, Huang, C.Y, Liu, P, Yin, Y, Liu, B, Caffrey, M, Melcher, K, Xu, Y, Wang, M.W, Xu, H.E, Jiang, Y. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | A common antagonistic mechanism for class A GPCRs revealed by the structure of the human 5-HT1B serotonin receptor bound to an antagonist
Cell Discov, 2018
|
|
2J0M
 
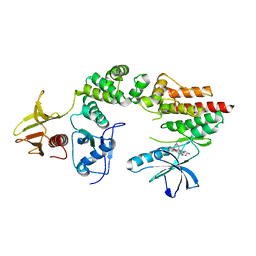 | | Crystal structure a two-chain complex between the FERM and kinase domains of focal adhesion kinase. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase.
Cell(Cambridge,Mass.), 129, 2007
|
|
2J0K
 
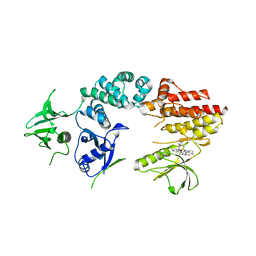 | | Crystal structure of a fragment of focal adhesion kinase containing the FERM and kinase domains. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase.
Cell(Cambridge,Mass.), 129, 2007
|
|
2J0J
 
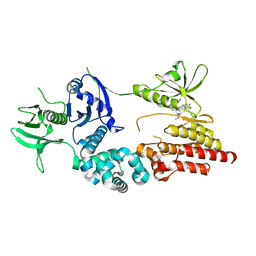 | | Crystal structure of a fragment of focal adhesion kinase containing the FERM and kinase domains. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase
Cell(Cambridge,Mass.), 129, 2007
|
|
2J0L
 
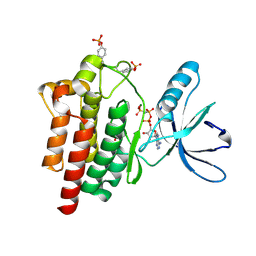 | | Crystal structure of a the active conformation of the kinase domain of focal adhesion kinase with a phosphorylated activation loop. | | Descriptor: | FOCAL ADHESION KINASE 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase
Cell(Cambridge,Mass.), 129, 2007
|
|
8WJY
 
 | | PKMYT1_Cocrystal_Cpd 4 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Wang, Y, Wang, C, Liu, T, Qi, H, Chen, S, Cai, X, Zhang, M, Aliper, A, Ren, F, Ding, X, Zhavoronkov, A. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of Tetrahydropyrazolopyrazine Derivatives as Potent and Selective MYT1 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
5XEZ
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
7D72
 
 | | Cryo-EM structures of human GMPPA/GMPPB complex bound to GDP-Mannose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
