3KBX
 
 | |
1S26
 
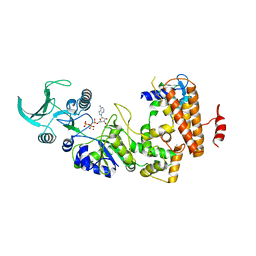 | | Structure of Anthrax Edema Factor-Calmodulin-alpha,beta-methyleneadenosine 5'-triphosphate Complex Reveals an Alternative Mode of ATP Binding to the Catalytic Site | | Descriptor: | CALCIUM ION, Calmodulin, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Bohm, A, Tang, W.-J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of anthrax edema factor-calmodulin-adenosine-5'-(alpha,beta-methylene)-triphosphate complex reveals an alternative mode of ATP binding to the catalytic site
Biochem.Biophys.Res.Commun., 317, 2004
|
|
1LVC
 
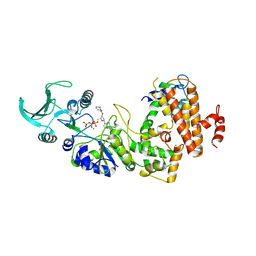 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin and 2' deoxy, 3' anthraniloyl ATP | | Descriptor: | 3'ANTHRANILOYL-2'-DEOXY-ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, YTTERBIUM (III) ION, ... | | Authors: | Shen, Y, Lee, Y.-S, Soelaiman, S, Bergson, P, Lu, D, Chen, A, Beckingham, K, Grabarek, Z, Mrksich, M, Tang, W.-J. | | Deposit date: | 2002-05-28 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Physiological calcium concentrations regulate calmodulin binding and catalysis of adenylyl cyclase exotoxins
Embo J., 21, 2002
|
|
1Y0V
 
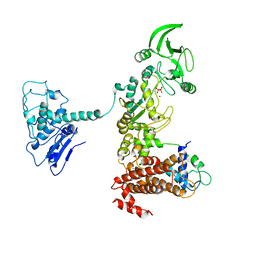 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and pyrophosphate | | Descriptor: | CALCIUM ION, Calmodulin, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.-J. | | Deposit date: | 2004-11-16 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor
Embo J., 24, 2005
|
|
2YPU
 
 | | human insulin degrading enzyme E111Q in complex with inhibitor compound 41367 | | Descriptor: | 2-[[2-[[(2S)-3-(3H-IMIDAZOL-4-YL)-1-METHOXY-1-OXO-PROPAN-2-YL]AMINO]-2-OXO-ETHYL]-(PHENYLMETHYL)AMINO]ETHANOIC ACID, INSULIN-DEGRADING ENZYME, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.-J. | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Imidazole-Derived 2-[N-Carbamoylmethyl-Alkylamino]Acetic Acids,Substrate-Dependent Modulators of Insulin-Degrading Enzyme in Amyloid-Beta Hydrolysis
Eur J Med Chem, 79C, 2014
|
|
1K93
 
 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, CALMODULIN-SENSITIVE ADENYLATE CYCLASE, ... | | Authors: | Drum, C.L, Yan, S.-Z, Bard, J, Shen, Y.-Q, Lu, D, Soelaiman, S, Grabarek, Z, Bohm, A, Tang, W.-J. | | Deposit date: | 2001-10-26 | | Release date: | 2002-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin.
Nature, 415, 2002
|
|
1K90
 
 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) in complex with calmodulin and 3' deoxy-ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CALMODULIN, ... | | Authors: | Drum, C.L, Yan, S.-Z, Bard, J, Shen, Y.-Q, Lu, D, Soelaiman, S, Grabarek, Z, Bohm, A, Tang, W.-J. | | Deposit date: | 2001-10-26 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin.
Nature, 415, 2002
|
|
1K8T
 
 | | Crystal structure of the adenylyl cyclase domain of anthrax edema factor (EF) | | Descriptor: | CALMODULIN-SENSITIVE ADENYLATE CYCLASE, NICKEL (II) ION, SULFATE ION | | Authors: | Drum, C.L, Yan, S.-Z, Bard, J, Shen, Y.-Q, Lu, D, Soelaiman, S, Grabarek, Z, Bohm, A, Tang, W.-J. | | Deposit date: | 2001-10-25 | | Release date: | 2002-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin
Nature, 415, 2002
|
|
3N57
 
 | |
3N56
 
 | |
5L2U
 
 | |
4MHE
 
 | | Crystal structure of CC-chemokine 18 | | Descriptor: | ACETATE ION, C-C motif chemokine 18 | | Authors: | Liang, W.G, Tang, W.-J. | | Deposit date: | 2013-08-29 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
3E50
 
 | | Crystal structure of human insulin degrading enzyme in complex with transforming growth factor-alpha | | Descriptor: | Insulin-degrading enzyme, Protransforming growth factor alpha, ZINC ION | | Authors: | Guo, Q, Manolopoulou, M, Tang, W.-J. | | Deposit date: | 2008-08-12 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for the Recognition and Cleavages of IGF-II, TGF-alpha, and Amylin by Human Insulin-Degrading Enzyme.
J.Mol.Biol., 395, 2010
|
|
3E4Z
 
 | | Crystal structure of human insulin degrading enzyme in complex with insulin-like growth factor II | | Descriptor: | Insulin-degrading enzyme, Insulin-like growth factor II, ZINC ION | | Authors: | Guo, Q, Manolopoulou, M, Tang, W.-J. | | Deposit date: | 2008-08-12 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Molecular Basis for the Recognition and Cleavages of IGF-II, TGF-alpha, and Amylin by Human Insulin-Degrading Enzyme.
J.Mol.Biol., 395, 2010
|
|
1PK0
 
 | | Crystal Structure of the EF3-CaM complexed with PMEApp | | Descriptor: | (ADENIN-9-YL-ETHOXYMETHYL)-HYDROXYPHOSPHINYL-DIPHOSPHATE, CALCIUM ION, Calmodulin, ... | | Authors: | Shen, Y, Tang, W.J. | | Deposit date: | 2003-06-04 | | Release date: | 2004-02-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Selective inhibition of anthrax edema factor by adefovir, a drug for chronic hepatitis B virus infection.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2R7I
 
 | | Crystal structure of catalytic subunit of protein kinase CK2 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shen, Y. | | Deposit date: | 2007-09-08 | | Release date: | 2008-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Crystal structures of catalytic and regulatory subunits of rat protein kinase CK2
CHIN.SCI.BULL., 54, 2009
|
|
2R6M
 
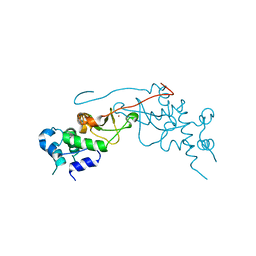 | | Crystal structure of rat CK2-beta subunit | | Descriptor: | Casein kinase II subunit beta, ZINC ION | | Authors: | Shen, Y. | | Deposit date: | 2007-09-06 | | Release date: | 2008-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of catalytic and regulatory subunits of rat protein kinase CK2
CHIN.SCI.BULL., 54, 2009
|
|
3OFI
 
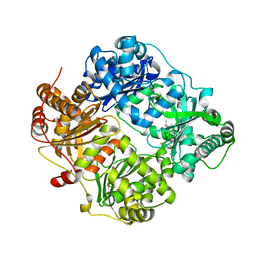 | | Crystal structure of human insulin-degrading enzyme in complex with ubiquitin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Insulin-degrading enzyme, Ubiquitin, ... | | Authors: | Kalas, V, Ralat, L.A, Tang, W.-J. | | Deposit date: | 2010-08-15 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Ubiquitin is a novel substrate for human insulin-degrading enzyme.
J.Mol.Biol., 406, 2011
|
|
2G47
 
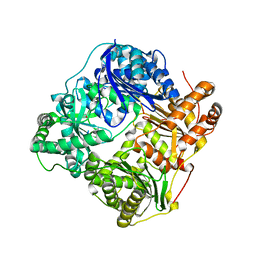 | |
2G48
 
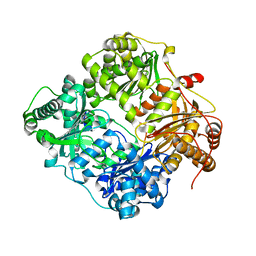 | |
2G54
 
 | |
2G56
 
 | |
2G49
 
 | |
5CMD
 
 | |
