2KBS
 
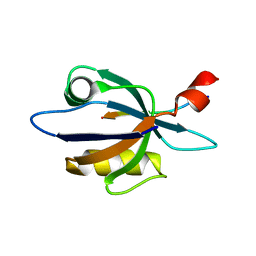 | | Solution structure of harmonin PDZ2 in complex with the carboxyl tail peptide of cadherin23 | | Descriptor: | Harmonin, octameric peptide from Cadherin-23 | | Authors: | Pan, L, Yan, J, Wu, L, Zhang, M. | | Deposit date: | 2008-12-05 | | Release date: | 2009-03-31 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Assembling stable hair cell tip link complex via multidentate interactions between harmonin and cadherin 23
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KBR
 
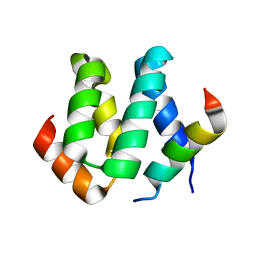 | | Solution structure of harmonin N terminal domain in complex with a internal peptide of cadherin23 | | Descriptor: | 18-meric peptide from Cadherin-23, Harmonin | | Authors: | Pan, L, Yan, J, Wu, L, Zhang, M. | | Deposit date: | 2008-12-05 | | Release date: | 2009-03-31 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Assembling stable hair cell tip link complex via multidentate interactions between harmonin and cadherin 23
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2PKU
 
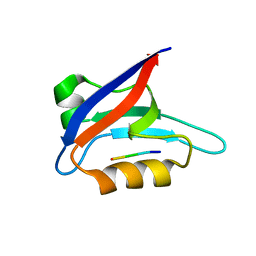 | | Solution structure of PICK1 PDZ in complex with the carboxyl tail peptide of GluR2 | | Descriptor: | PRKCA-binding protein, peptide (GLU)(SER)(VAL)(LYS)(ILE) | | Authors: | Pan, L, Wu, H, Shen, C, Shi, Y, Xia, J, Zhang, M. | | Deposit date: | 2007-04-18 | | Release date: | 2007-11-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Clustering and synaptic targeting of PICK1 requires direct interaction between the PDZ domain and lipid membranes
Embo J., 26, 2007
|
|
6NHW
 
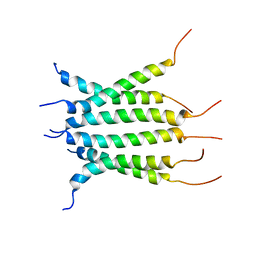 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6NHY
 
 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
2L7T
 
 | |
2KBQ
 
 | |
2LSR
 
 | | Solution structure of harmonin N terminal domain in complex with a exon68 encoded peptide of cadherin23 | | Descriptor: | Harmonin, peptide from Cadherin-23 | | Authors: | Pan, L, Wu, L, Zhang, C, Zhang, M. | | Deposit date: | 2012-05-04 | | Release date: | 2012-08-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Large protein assemblies formed by multivalent interactions between cadherin23 and harmonin suggest a stable anchorage structure at the tip link of stereocilia.
J.Biol.Chem., 287, 2012
|
|
5BTU
 
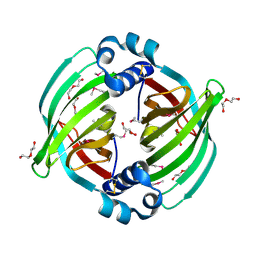 | |
5BU3
 
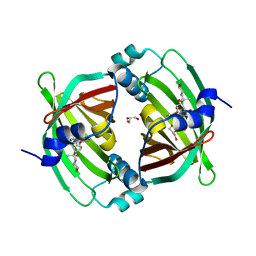 | | Crystal Structure of Diels-Alderase PyrI4 in complex with its product | | Descriptor: | (4S,4aS,6aS,8R,9R,10aR,13R,14aS,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,5,6,6a,7,8,9,10,10a,13,14,18a,18b-hexadecahydro-1H-14a,17-(metheno)benzo[b]naphtho[2,1-h]azacyclododecine-16,18(15H,17H)-dione, GLYCEROL, PyrI4 | | Authors: | Pan, L, Guo, Y, Liu, J. | | Deposit date: | 2015-06-03 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Enzyme-Dependent [4 + 2] Cycloaddition Depends on Lid-like Interaction of the N-Terminal Sequence with the Catalytic Core in PyrI4
Cell Chem Biol, 23, 2016
|
|
3K1R
 
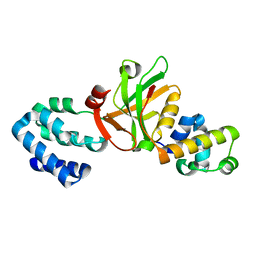 | |
2MXP
 
 | |
3SHU
 
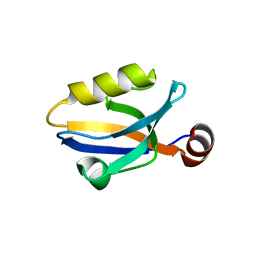 | | Crystal structure of ZO-1 PDZ3 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Yu, J, Pan, L, Chen, J, Yu, H, Zhang, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of the PDZ3-SH3-GuK Tandem of ZO-1 Suggests a Supramodular Organization of the Conserved MAGUK Family Scaffold Core
To be Published
|
|
3SHW
 
 | | Crystal structure of ZO-1 PDZ3-SH3-Guk supramodule complex with Connexin-45 peptide | | Descriptor: | Gap junction gamma-1 protein, Tight junction protein ZO-1 | | Authors: | Yu, J, Pan, L, Chen, J, Yu, H, Zhang, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of the PDZ3-SH3-GuK Tandem of ZO-1 Suggests a Supramodular Organization of the Conserved MAGUK Family Scaffold Core
To be Published
|
|
3CYY
 
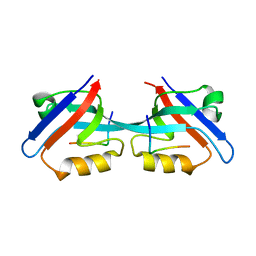 | |
5F5N
 
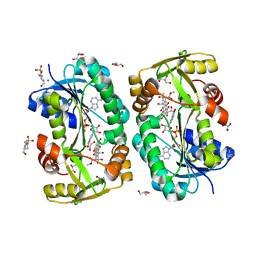 | |
5F5L
 
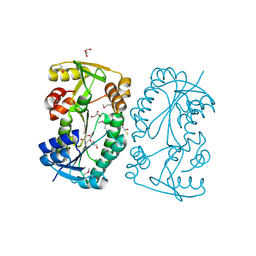 | |
5XGV
 
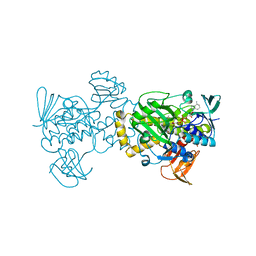 | |
5YT6
 
 | |
7X9Q
 
 | | Crystal structure of human STING complexed with compound BSP16 | | Descriptor: | (2R)-4-(5,6-dimethoxy-1-benzoselenophen-2-yl)-2-ethyl-4-oxidanylidene-butanoic acid, Stimulator of interferon genes protein | | Authors: | Pan, L, Guan, X, Feng, X, Li, Z, Bian, J. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Selenium-Containing STING Agonists as Orally Available Antitumor Agents.
J.Med.Chem., 65, 2022
|
|
7X9P
 
 | | Crystal structure of human STING complexed with compound BSP17 | | Descriptor: | 4-[6-methoxy-5-[3-[[6-methoxy-2-(4-oxidanyl-4-oxidanylidene-butanoyl)-1-benzoselenophen-5-yl]oxy]propoxy]-1-benzoselenophen-2-yl]-4-oxidanylidene-butanoic acid, Stimulator of interferon genes protein | | Authors: | Pan, L, Guan, X, Feng, X, Li, Z, Bian, J. | | Deposit date: | 2022-03-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Selenium-containing STING agonists as orally available anti tumor agents
To be published
|
|
5GP7
 
 | | Structural basis for the binding between Tankyrase-1 and USP25 | | Descriptor: | GLYCEROL, Tankyrase-1, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Liu, J, Xu, D, Fu, T, Pan, L. | | Deposit date: | 2016-08-01 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | USP25 regulates Wnt signaling by controlling the stability of tankyrases
Genes Dev., 31, 2017
|
|
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | Descriptor: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | Authors: | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
4XKL
 
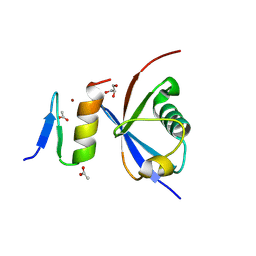 | | Crystal structure of NDP52 ZF2 in complex with mono-ubiquitin | | Descriptor: | ACETATE ION, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | Authors: | Xie, X, Li, F, Wang, Y, Lin, Z, Chen, X, Liu, J, Pan, L. | | Deposit date: | 2015-01-12 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2
Autophagy, 11, 2015
|
|
7V8H
 
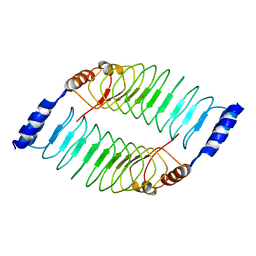 | | Crystal structure of LRR domain from Shigella flexneri IpaH1.4 | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Liu, J, Wang, Y, Pan, L. | | Deposit date: | 2021-08-23 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanistic insights into the subversion of the linear ubiquitin chain assembly complex by the E3 ligase IpaH1.4 of Shigella flexneri.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
