4M36
 
 | |
4M37
 
 | |
4Y33
 
 | | Crystal of NO66 in complex with Ni(II)and N-oxalylglycine (NOG) | | Descriptor: | Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, N-OXALYLGLYCINE, NICKEL (II) ION | | Authors: | Wang, C, Zhang, Q, Zang, J. | | Deposit date: | 2015-02-10 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Y3O
 
 | | Crystal structure of Ribosomal oxygenase NO66 in complex with substrate Rpl8 peptide and Ni(II) and cofactor N-oxalyglycine | | Descriptor: | ACETATE ION, Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, GLYCEROL, ... | | Authors: | Wang, C, Zhang, Q, Zang, J. | | Deposit date: | 2015-02-10 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2LGK
 
 | | NMR Structure of UHRF1 PHD domains in a complex with histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
7ZHI
 
 | |
7ZP2
 
 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum in complex with BDA-04 | | Descriptor: | Aspartate carbamoyltransferase, SODIUM ION, SULFATE ION, ... | | Authors: | Wang, C, Zhang, B, Groves, M.R. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Discovery of Small-Molecule Allosteric Inhibitors of Pf ATC as Antimalarials.
J.Am.Chem.Soc., 144, 2022
|
|
7ZGS
 
 | |
7ZST
 
 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum in complex with FLA-01 | | Descriptor: | 2-azanyl-~{N}-(2-methoxyethyl)-5-phenyl-thiophene-3-carboxamide, Aspartate carbamoyltransferase, GLYCEROL, ... | | Authors: | Wang, C, Zhang, B, Groves, M.R. | | Deposit date: | 2022-05-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Small-Molecule Allosteric Inhibitors of Pf ATC as Antimalarials.
J.Am.Chem.Soc., 144, 2022
|
|
7ZID
 
 | |
8EFP
 
 | |
8ET1
 
 | |
8ET2
 
 | | CryoEM structure of the GSDMB pore | | Descriptor: | Isoform 1 of Gasdermin-B | | Authors: | Wang, C, Ruan, J. | | Deposit date: | 2022-10-15 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.96 Å) | | Cite: | Structural basis for GSDMB pore formation and its targeting by IpaH7.8.
Nature, 616, 2023
|
|
4EKZ
 
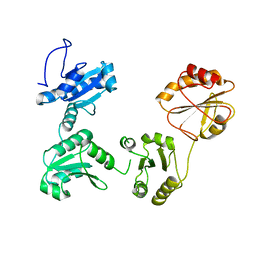 | | Crystal structure of reduced hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
4EL1
 
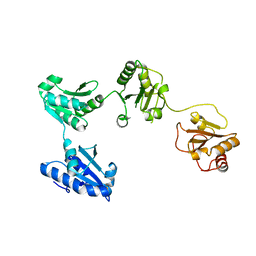 | | Crystal structure of oxidized hPDI (abb'xa') | | Descriptor: | Protein disulfide-isomerase | | Authors: | Wang, C, Li, W, Ren, J, Ke, H, Gong, W, Feng, W, Wang, C.-C. | | Deposit date: | 2012-04-10 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase
Antioxid Redox Signal, 19, 2013
|
|
5E8H
 
 | | Crystal structure of geranylfarnesyl pyrophosphate synthases 2 from Arabidopsis thaliana | | Descriptor: | Geranylgeranyl pyrophosphate synthase 3, chloroplastic | | Authors: | Wang, C, Chen, Q, Wang, G, Zhang, P. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-11 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
5E8K
 
 | | Crystal structure of polyprenyl pyrophosphate synthase 2 from Arabidopsis thaliana | | Descriptor: | Geranylgeranyl pyrophosphate synthase 10, mitochondrial | | Authors: | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.028 Å) | | Cite: | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
4JPZ
 
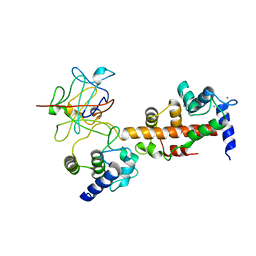 | | Voltage-gated sodium channel 1.2 C-terminal domain in complex with FGF13U and Ca2+/calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Fibroblast growth factor 13, ... | | Authors: | Wang, C, Chung, B.C, Yan, H, Wang, H.G, Lee, S.Y, Pitt, G.S. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural analyses of Ca(2+)/CaM interaction with NaV channel C-termini reveal mechanisms of calcium-dependent regulation.
Nat Commun, 5, 2014
|
|
5E8L
 
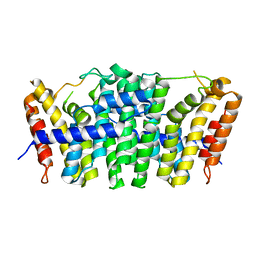 | | Crystal structure of geranylgeranyl pyrophosphate synthase 11 from Arabidopsis thaliana | | Descriptor: | Heterodimeric geranylgeranyl pyrophosphate synthase large subunit 1, chloroplastic | | Authors: | Wang, C, Chen, Q, Fan, D, Li, J, Wang, G, Zhang, P. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants.
Mol Plant, 9, 2016
|
|
4JQ0
 
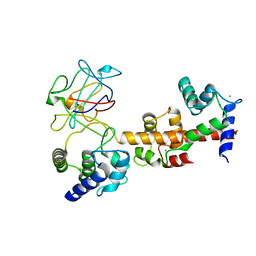 | | Voltage-gated sodium channel 1.5 C-terminal domain in complex with FGF12B and Ca2+/calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Fibroblast growth factor 12, ... | | Authors: | Wang, C, Chung, B.C, Yan, H, Wang, H.G, Lee, S.Y, Pitt, G.S. | | Deposit date: | 2013-03-19 | | Release date: | 2014-05-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Structural analyses of Ca(2+)/CaM interaction with NaV channel C-termini reveal mechanisms of calcium-dependent regulation.
Nat Commun, 5, 2014
|
|
7UVP
 
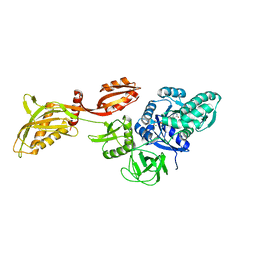 | |
1F0Z
 
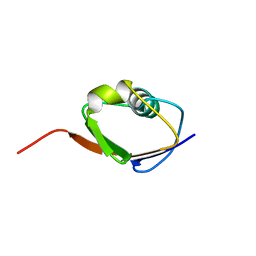 | | SOLUTION STRUCTURE OF THIS, THE SULFUR CARRIER PROTEIN IN E.COLI THIAMIN BIOSYNTHESIS | | Descriptor: | THIS PROTEIN | | Authors: | Wang, C, Xi, J, Begley, T.P, Nicholson, L.K. | | Deposit date: | 2000-05-17 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ThiS and implications for the evolutionary roots of ubiquitin.
Nat.Struct.Biol., 8, 2001
|
|
7D7N
 
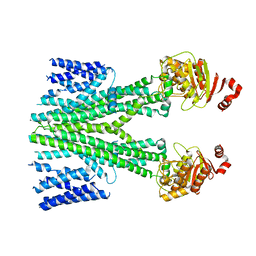 | | Cryo-EM structure of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
7D7R
 
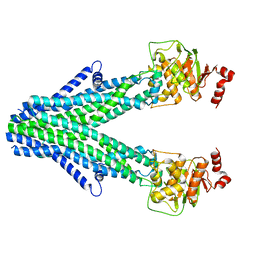 | | Cryo-EM structure of the core domain of human ABCB6 transporter | | Descriptor: | ATP-binding cassette sub-family B member 6, mitochondrial | | Authors: | Wang, C, Cao, C, Wang, N, Wang, X, Zhang, X.C. | | Deposit date: | 2020-10-05 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-electron microscopy structure of human ABCB6 transporter.
Protein Sci., 29, 2020
|
|
4M38
 
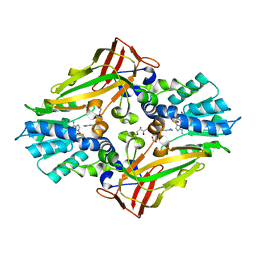 | |
