3CIA
 
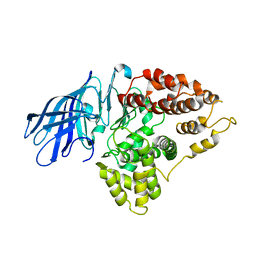 | | Crystal structure of cold-aminopeptidase from Colwellia psychrerythraea | | Descriptor: | ZINC ION, cold-active aminopeptidase | | Authors: | Bauvois, C, Jacquamet, L, Borel, F, Ferrer, J.-L. | | Deposit date: | 2008-03-11 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Cold-active Aminopeptidase from Colwellia psychrerythraea, a Close Structural Homologue of the Human Bifunctional Leukotriene A4 Hydrolase.
J.Biol.Chem., 283, 2008
|
|
2R59
 
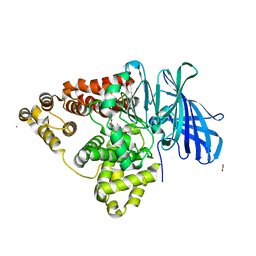 | | Leukotriene A4 hydrolase complexed with inhibitor RB3041 | | Descriptor: | ACETIC ACID, Leukotriene A-4 hydrolase, N-{(2S)-3-[(R)-[(1R)-1-amino-2-phenylethyl](hydroxy)phosphoryl]-2-benzylpropanoyl}-L-phenylalanine, ... | | Authors: | Tholander, F, Haeggstrom, J.Z, Thunnissen, M, Muroya, A, Roques, B.P, Fournie-Zaluski, M.C. | | Deposit date: | 2007-09-03 | | Release date: | 2008-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
7PFS
 
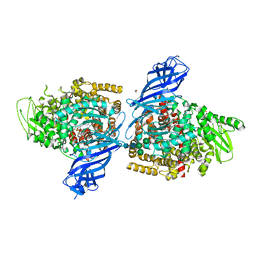 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide ((1R)-1-Amino-3-phenylpropyl){2-([1,1:3,1-terphenyl]-5-ylmethyl)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)-amino]-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
6YDX
 
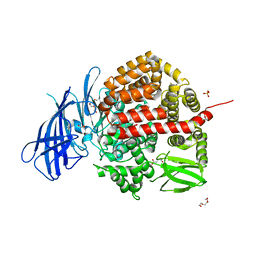 | | Insulin-regulated aminopeptidase complexed with a macrocyclic peptidic inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-[[[(4~{R},8~{S},11~{S})-11-azanyl-8-[(4-hydroxyphenyl)methyl]-6,10-bis(oxidanylidene)-1,2-dithia-5,9-diazacyclotridec-4-yl]carbonylamino]methyl]phenyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Saridakis, E, Giastas, P, Stratikos, E. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Inhibition of Insulin-Regulated Aminopeptidase by a Macrocyclic Peptidic Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
6RQX
 
 | |
2GTQ
 
 | | Crystal structure of aminopeptidase N from human pathogen Neisseria meningitidis | | Descriptor: | SULFATE ION, ZINC ION, aminopeptidase N | | Authors: | Nocek, B, Mulligan, R, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of aminopeptidase N from human pathogen Neisseria meningitidis.
Proteins, 70, 2007
|
|
7SH0
 
 | | CRYSTAL STRUCTURE OF ENDOPLASMIC RETICULUM AMINOPEPTIDASE 2 (ERAP2) COMPLEX WITH A HIGHLY SELECTIVE AND POTENT SMALL MOLECULE | | Descriptor: | (2S)-N-hydroxy-3-(4-methoxyphenyl)-2-[4-({[5-(pyridin-2-yl)thiophene-2-sulfonyl]amino}methyl)-1H-1,2,3-triazol-1-yl]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Li, L, Bouvier, M. | | Deposit date: | 2021-10-07 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of the First Selective Nanomolar Inhibitors of ERAP2 by Kinetic Target-Guided Synthesis.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7ZYF
 
 | | Insulin regulated aminopeptidase (IRAP) in complex with a nanomolar alpha hydroxy beta amino acid based inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Stratikos, E, Giastas, P, Papakyriakou, A. | | Deposit date: | 2022-05-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of Selective Nanomolar Inhibitors for Insulin-Regulated Aminopeptidase Based on alpha-Hydroxy-beta-amino Acid Derivatives of Bestatin.
J.Med.Chem., 65, 2022
|
|
6M8P
 
 | | Human ERAP1 bound to phosphinic pseudotripeptide inhibitor DG013 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 1, Nalpha-[(2S)-2-{[[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]methyl}-4-methylpentanoyl]-L-tryptophanamide, ... | | Authors: | Maben, Z, Stern, L.J. | | Deposit date: | 2018-08-22 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Conformational dynamics linked to domain closure and substrate binding explain the ERAP1 allosteric regulation mechanism.
Nat Commun, 12, 2021
|
|
5FWQ
 
 | | Apo structure of human Leukotriene A4 hydrolase | | Descriptor: | ACETATE ION, HUMAN LEUKOTRIENE A4 HYDROLASE, IMIDAZOLE, ... | | Authors: | Wittmann, S.K, Kalinowsky, L, Kramer, J, Bloecher, R, Steinhilber, D, Pogoryelov, D, Proschak, E, Heering, J. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Thermodynamic properties of leukotriene A4hydrolase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
8JKT
 
 | |
3KED
 
 | | Crystal structure of Aminopeptidase N in complex with 2,4-diaminobutyric acid | | Descriptor: | 2,4-DIAMINOBUTYRIC ACID, Aminopeptidase N, GLYCEROL, ... | | Authors: | Addlagatta, A, Gumpena, R. | | Deposit date: | 2009-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of alpha, beta- and alpha, gamma-Diamino Acid Scaffolds for the Inhibition of M1 Family Aminopeptidases
Chemmedchem, 6, 2011
|
|
7U0L
 
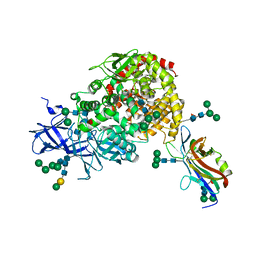 | | Crystal structure of the CCoV-HuPn-2018 RBD (domain B) in complex with canine APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
1SQM
 
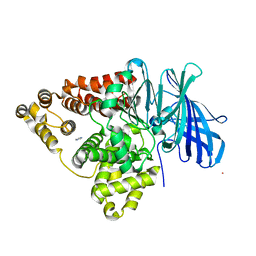 | | STRUCTURE OF [R563A] LEUKOTRIENE A4 HYDROLASE | | Descriptor: | ACETIC ACID, IMIDAZOLE, LEUKOTRIENE A-4 HYDROLASE, ... | | Authors: | Tholander, F.O.T, Rudberg, P.C, Thunnissen, M.M.G.M, Haeggstrom, J.Z. | | Deposit date: | 2004-03-19 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Leukotriene A4 hydrolase: identification of a common carboxylate recognition site for the epoxide hydrolase and aminopeptidase substrates
J.Biol.Chem., 279, 2004
|
|
6O5H
 
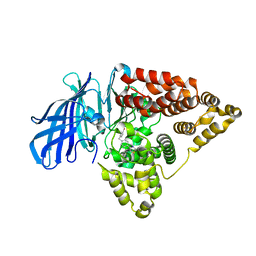 | | The effect of modifier structure on the activation of leukotriene A4 hydrolase aminopeptidase activity. | | Descriptor: | 4-{4-[(4-methoxyphenyl)methyl]phenyl}-1,3-thiazol-2-amine, Leukotriene A-4 hydrolase, ZINC ION | | Authors: | Noble, S.M, Lee, K.H, Paige, M. | | Deposit date: | 2019-03-03 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Effect of Modifier Structure on the Activation of Leukotriene A4Hydrolase Aminopeptidase Activity.
J.Med.Chem., 62, 2019
|
|
5J6S
 
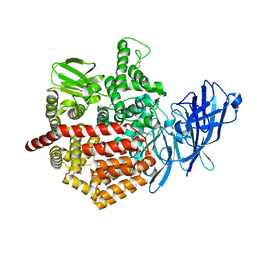 | | Crystal structure of Endoplasmic Reticulum Aminopeptidase 2 (ERAP2) in complex with a hydroxamic derivative ligand | | Descriptor: | (2S)-N~1~-benzyl-2-[(4-fluorophenyl)methyl]-N~3~-hydroxypropanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saridakis, E, Giastas, P, Mpakali, A, Deprez-Poulain, R, Stratikos, E. | | Deposit date: | 2016-04-05 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
6MGQ
 
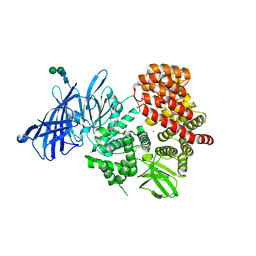 | | ERAP1 in the open conformation bound to 10mer phosphinic inhibitor DG014 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 1, Phosphinic inhibitor DG014, ... | | Authors: | Stern, L.J, Maben, Z. | | Deposit date: | 2018-09-14 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Conformational dynamics linked to domain closure and substrate binding explain the ERAP1 allosteric regulation mechanism.
Nat Commun, 12, 2021
|
|
2DQ6
 
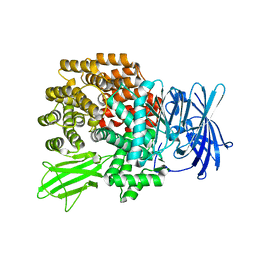 | | Crystal Structure of Aminopeptidase N from Escherichia coli | | Descriptor: | Aminopeptidase N, SULFATE ION, ZINC ION | | Authors: | Nakajima, Y, Onohara, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
8AVA
 
 | |
5K1V
 
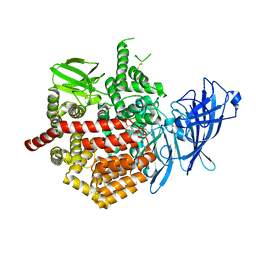 | | Crystal structure of Endoplasmic Reticulum aminopeptidase 2 (ERAP2) in complex with a diaminobenzoic acid derivative ligand. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Papakyriakou, A, Giastas, P, Mpakali, A, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
8AWH
 
 | |
7KZE
 
 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | Descriptor: | 1-benzyl-4-methoxybenzene, Leukotriene A-4 hydrolase, TRIETHYLENE GLYCOL, ... | | Authors: | Lee, K.H, Shim, Y, Paige, M, Noble, S.M. | | Deposit date: | 2020-12-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
1Z5H
 
 | | Crystal structures of the Tricorn interacting Factor F3 from Thermoplasma acidophilum | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-18 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 349, 2005
|
|
7LLQ
 
 | | Substrate-dependent divergence of leukotriene A4 hydrolase aminopeptidase activity | | Descriptor: | 1-benzyl-4-methoxybenzene, 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, Leukotriene A-4 hydrolase, ... | | Authors: | Lee, K.H, Lee, S.H, Shim, Y.M, Paige, M, Noble, S.M. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Substrate-dependent modulation of the leukotriene A 4 hydrolase aminopeptidase activity and effect in a murine model of acute lung inflammation.
Sci Rep, 12, 2022
|
|
6Q4R
 
 | | High-resolution crystal structure of ERAP1 with bound phosphinic transition-state analogue inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Neu, M, Rowland, P, Stratikos, E. | | Deposit date: | 2018-12-06 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-Resolution Crystal Structure of Endoplasmic Reticulum Aminopeptidase 1 with Bound Phosphinic Transition-State Analogue Inhibitor.
Acs Med.Chem.Lett., 10, 2019
|
|
