6NUC
 
 | | Structure of Calcineurin in complex with NHE1 peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the binding and selective dephosphorylation of Na+/H+exchanger 1 by calcineurin.
Nat Commun, 10, 2019
|
|
6NUF
 
 | | Structure of Calcineurin in complex with NHE1 peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the binding and selective dephosphorylation of Na+/H+exchanger 1 by calcineurin.
Nat Commun, 10, 2019
|
|
4BVN
 
 | | Ultra-thermostable beta1-adrenoceptor with cyanopindolol bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, ... | | Authors: | Miller, J, Nehme, R, Warne, T, Edwards, P.C, Leslie, A.G.W, Schertler, G, Tate, C.G. | | Deposit date: | 2013-06-26 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A Resolution Structure of Cyanopindolol-Bound Beta1- Adrenoceptor Identifies an Intramembrane Na+ Ion that Stabilises the Ligand-Free Receptor.
Plos One, 9, 2014
|
|
2KBV
 
 | | Structural and functional analysis of TM XI of the NHE1 isoform of thE NA+/H+ exchanger | | Descriptor: | Sodium/hydrogen exchanger 1 | | Authors: | Lee, B.L, Li, X, Liu, Y, Sykes, B.D, Fliegel, L. | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Analysis of Transmembrane XI of the NHE1 Isoform of the Na+/H+ Exchanger
J.Biol.Chem., 284, 2009
|
|
4NYP
 
 | |
6A31
 
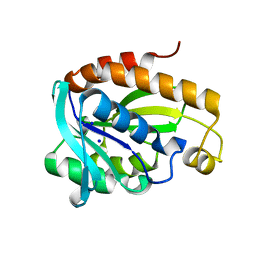 | | Crystal structure of Na+ bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 2.19 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase, SODIUM ION | | Authors: | Bairagya, H.R, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2018-06-14 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of Na+ bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 2.19 A resolution
To Be Published
|
|
2K3C
 
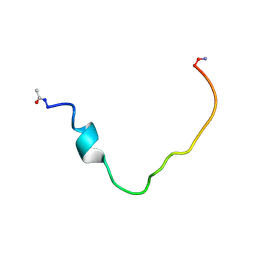 | | Structural and Functional Characterization of TM IX of the NHE1 Isoform of the Na+/H+ Exchanger | | Descriptor: | TMIX peptide | | Authors: | Reddy, T, Ding, J, Li, X, Sykes, B.D, Fliegel, L, Rainey, J.K. | | Deposit date: | 2008-05-01 | | Release date: | 2008-06-03 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of Transmembrane Segment IX of the NHE1 Isoform of the Na+/H+ Exchanger.
J.Biol.Chem., 283, 2008
|
|
2L0E
 
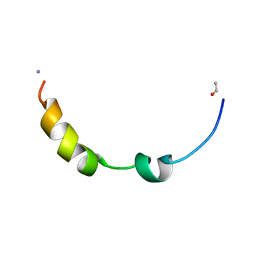 | |
2MKV
 
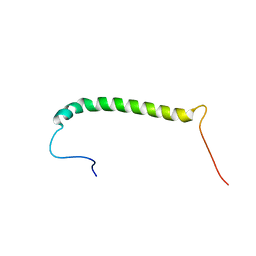 | |
4M64
 
 | |
3R3G
 
 | | Structure of human thrombin with residues 145-150 of murine thrombin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Thrombin Heavy Chain, ... | | Authors: | Pozzi, N, Chen, R, Chen, Z, Bah, A, Di Cera, E. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rigidification of the autolysis loop enhances Na(+) binding to thrombin.
Biophys.Chem., 159, 2011
|
|
4NUN
 
 | | Crystal structure of copper-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, COPPER (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
4NUO
 
 | | Crystal structure of zinc-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, ZINC ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
4NUK
 
 | | Crystal structure of nickel-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, NICKEL (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
4NUI
 
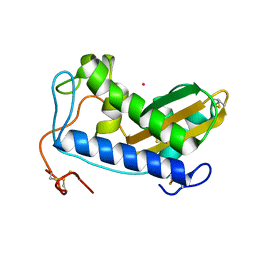 | | Crystal structure of cobalt-bound Na-ASP-2 | | Descriptor: | Ancylostoma secreted protein 2, COBALT (II) ION | | Authors: | Hofmann, A. | | Deposit date: | 2013-12-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the equatorial groove of the hookworm protein and vaccine candidate antigen, Na-ASP-2.
Int.J.Biochem.Cell Biol., 50, 2014
|
|
2JO1
 
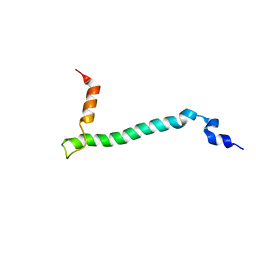 | | Structure of the Na,K-ATPase regulatory protein FXYD1 in micelles | | Descriptor: | Phospholemman | | Authors: | Teriete, P, Franzin, C.M, Choi, J, Marassi, F.M. | | Deposit date: | 2007-02-18 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the Na,K-ATPase regulatory protein FXYD1 in micelles
Biochemistry, 46, 2007
|
|
3F6U
 
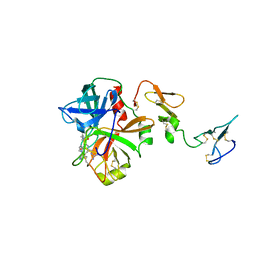 | | Crystal structure of human Activated Protein C (APC) complexed with PPACK | | Descriptor: | CALCIUM ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, ... | | Authors: | Schmidt, A.E, Padmanabhan, K, Underwood, M.C, Bode, W, Mather, T, Bajaj, S.P. | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermodynamic linkage between the S1 site, the Na+ site, and the Ca2+ site in the protease domain of human activated protein C (APC).
J.Biol.Chem., 277, 2002
|
|
6J8E
 
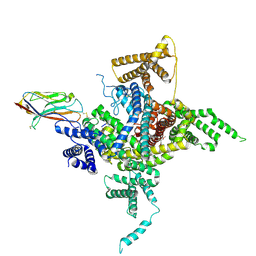 | | Human Nav1.2-beta2-KIIIA ternary complex | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, X, Li, Z, Huang, X, Huang, G, Yan, N. | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for pore blockade of human Na+channel Nav1.2 by the mu-conotoxin KIIIA.
Science, 363, 2019
|
|
4LLH
 
 | | Substrate bound outward-open state of the symporter BetP | | Descriptor: | 2-(trimethyl-lambda~5~-arsanyl)ethanol, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, CHLORIDE ION, ... | | Authors: | Perez, C, Faust, B, Ziegler, C. | | Deposit date: | 2013-07-09 | | Release date: | 2014-05-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate-bound outward-open state of the betaine transporter BetP provides insights into Na(+) coupling.
Nat Commun, 5, 2014
|
|
3N23
 
 | | Crystal structure of the high affinity complex between ouabain and the E2P form of the sodium-potassium pump | | Descriptor: | MAGNESIUM ION, Na+/K+ ATPase gamma subunit transcript variant a, OUABAIN, ... | | Authors: | Yatime, L, Laursen, M, Morth, J.P, Esmann, M, Nissen, P, Fedosova, N.U. | | Deposit date: | 2010-05-17 | | Release date: | 2011-01-19 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Structural insights into the high affinity binding of cardiotonic steroids to the Na+,K+-ATPase.
J.Struct.Biol., 174, 2011
|
|
6BJF
 
 | | NMR Structural and biophysical functional analysis of intracellular loop 5 of the NHE1 isoform of the Na+/H+ exchanger. | | Descriptor: | GLY-LEU-THR-TRP-PHE-ILE-ASN-LYS-PHE-ARG-ILE-VAL-LYS | | Authors: | McKay, R, Wong, K, Towle, K, Fliegel, L. | | Deposit date: | 2017-11-06 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Diverse residues of intracellular loop 5 of the Na+/H+exchanger modulate proton sensing, expression, activity and targeting.
Biochim Biophys Acta Biomembr, 1861, 2019
|
|
4JN7
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Bouvier, J.T, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE
To be Published
|
|
4BEM
 
 | | Crystal structure of the F-type ATP synthase c-ring from Acetobacterium woodii. | | Descriptor: | ACETATE ION, F1FO ATPASE C1 SUBUNIT, F1FO ATPASE C2 SUBUNIT, ... | | Authors: | Matthies, D, Meier, T, Yildiz, O. | | Deposit date: | 2013-03-11 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Resolution Structure and Mechanism of an F/V-Hybrid Rotor Ring in a Na+-Coupled ATP Synthase
Nat.Commun., 5, 2014
|
|
3AOU
 
 | | Structure of the Na+ unbound rotor ring modified with N,N f-Dicyclohexylcarbodiimide of the Na+-transporting V-ATPase | | Descriptor: | DICYCLOHEXYLUREA, UNDECYL-MALTOSIDE, V-type sodium ATPase subunit K | | Authors: | Mizutani, K, Yamamoto, M, Yamato, I, Kakinuma, Y, Shirouzu, M, Yokoyama, S, Iwata, S, Murata, T. | | Deposit date: | 2010-10-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4DW8
 
 | | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Na crystal form I | | Descriptor: | Haloacid dehalogenase-like hydrolase, SODIUM ION, UNKNOWN LIGAND | | Authors: | Vetting, M.W, Wasserman, S.R, Morisco, L.L, Sojitra, S, Allen, K.N, Dunaway-Mariano, D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-24 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase (Target EFI-900331) from Bacteroides thetaiotaomicron with bound Na crystal form I
To be Published
|
|
