8T1M
 
 | | Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Agdanowski, M.P, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8T0B
 
 | |
8T1N
 
 | | Micro-ED Structure of a Novel Domain of Unknown Function Solved with AlphaFold | | Descriptor: | DUF1842 domain-containing protein | | Authors: | Miller, J.E, Cascio, D, Sawaya, M.R, Cannon, K.A, Rodriguez, J.A, Yeates, T.O. | | Deposit date: | 2023-06-02 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | AlphaFold-assisted structure determination of a bacterial protein of unknown function using X-ray and electron crystallography.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
4BVN
 
 | | Ultra-thermostable beta1-adrenoceptor with cyanopindolol bound | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, ... | | Authors: | Miller, J, Nehme, R, Warne, T, Edwards, P.C, Leslie, A.G.W, Schertler, G, Tate, C.G. | | Deposit date: | 2013-06-26 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A Resolution Structure of Cyanopindolol-Bound Beta1- Adrenoceptor Identifies an Intramembrane Na+ Ion that Stabilises the Ligand-Free Receptor.
Plos One, 9, 2014
|
|
4R7Z
 
 | | PfMCM-AAA double-octamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 21, MAGNESIUM ION | | Authors: | Miller, J.M, Arachea, B.T, Epling, L.B, Enemark, E.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Analysis of the crystal structure of an active MCM hexamer.
Elife, 3, 2014
|
|
4R7Y
 
 | | Crystal structure of an active MCM hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Miller, J.M, Arachea, B.T, Epling, L.B, Enemark, E.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of the crystal structure of an active MCM hexamer.
Elife, 3, 2014
|
|
2IOU
 
 | |
7L0B
 
 | | Crystal structure of hydroxyacyl glutathione hydrolase (GloB) from Staphylococcus aureus, apoenzyme | | Descriptor: | Hydroxyacylglutathione hydrolase, SULFATE ION, ZINC ION | | Authors: | Miller, J.J, Jez, J.M, Odom John, A.R. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided microbial targeting of antistaphylococcal prodrugs.
Elife, 10, 2021
|
|
7L0A
 
 | |
1JK1
 
 | | Zif268 D20A Mutant Bound to WT DNA Site | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*G)-3', 5'-D(*TP*CP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3', ZIF268, ... | | Authors: | Miller, J.C, Pabo, C.O. | | Deposit date: | 2001-07-11 | | Release date: | 2001-10-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition.
J.Mol.Biol., 313, 2001
|
|
1JK2
 
 | | Zif268 D20A mutant bound to the GCT DNA site | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*TP*G)-3', 5'-D(*TP*CP*AP*GP*CP*CP*CP*AP*CP*GP*C)-3', ZIF268, ... | | Authors: | Miller, J.C, Pabo, C.O. | | Deposit date: | 2001-07-11 | | Release date: | 2001-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition.
J.Mol.Biol., 313, 2001
|
|
1R1C
 
 | | PSEUDOMONAS AERUGINOSA W48F/Y72F/H83Q/Y108W-AZURIN RE(PHEN)(CO)3(HIS107) | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (I) ION | | Authors: | Miller, J.E, Gradinaru, C, Crane, B.R, Di Bilio, A.J. | | Deposit date: | 2003-09-23 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spectroscopy and reactivity of a photogenerated tryptophan radical in a structurally defined protein environment
J.Am.Chem.Soc., 125, 2003
|
|
5NFZ
 
 | | TUBULIN-MTC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methoxy-5-(2,3,4-trimethoxyphenyl)cyclohepta-2,4,6-trien-1-one, CALCIUM ION, ... | | Authors: | Field, J.J, Pera, B, Estevez Gallego, J, Calvo, E, Rodriguez-Salarichs, J, Saez-Calvo, G, Zuwerra, D, Jordi, M, Prota, A.E, Menchon, G, Miller, J.H, Altmann, K.-H, Diaz, J.F. | | Deposit date: | 2017-03-16 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Zampanolide Binding to Tubulin Indicates Cross-Talk of Taxane Site with Colchicine and Nucleotide Sites.
J. Nat. Prod., 81, 2018
|
|
5NG1
 
 | | TUBULIN-MTC-zampanolide complex | | Descriptor: | (2Z,4E)-N-[(S)-[(1S,2E,5S,8E,10Z,17S)-3,11-dimethyl-19-methylidene-7,13-dioxo-6,21-dioxabicyclo[15.3.1]henicosa-2,8,10-trien-5-yl](hydroxy)methyl]hexa-2,4-dienamide, (2~{Z},4~{E})-~{N}-[(~{S})-oxidanyl-[(1~{S},2~{E},5~{S},11~{R},17~{S},19~{R})-3,11,19-trimethyl-7,13-bis(oxidanylidene)-6,21-dioxabicyclo[15.3.1]henicos-2-en-5-yl]methyl]hexa-2,4-dienamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Field, J.J, Pera, B, Estevez Gallego, J, Calvo, E, Rodriguez-Salarichs, J, Saez-Calvo, G, Zuwerra, D, Jordi, M, Prota, A.E, Menchon, G, Miller, J.H, Altmann, K.-H, Diaz, J.F. | | Deposit date: | 2017-03-16 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zampanolide Binding to Tubulin Indicates Cross-Talk of Taxane Site with Colchicine and Nucleotide Sites.
J. Nat. Prod., 81, 2018
|
|
3UXM
 
 | | Structure Guided Development of Novel Thymidine Mimetics targeting Pseudomonas aeruginosa Thymidylate Kinase: from Hit to Lead Generation | | Descriptor: | 5'-deoxy-5'-fluorothymidine, MAGNESIUM ION, Thymidylate kinase | | Authors: | Choi, J.Y, Plummer, M.S, Starr, J, Desbonnet, C.R, Soutter, H.H, Chang, J, Miller, J.R, Dillman, K, Miller, A.A, Roush, W.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure guided development of novel thymidine mimetics targeting Pseudomonas aeruginosa thymidylate kinase: from hit to lead generation.
J.Med.Chem., 55, 2012
|
|
3UWK
 
 | | Structure Guided Development of Novel Thymidine Mimetics targeting Pseudomonas aeruginosa Thymidylate Kinase: from Hit to Lead Generation | | Descriptor: | 1-methyl-6-phenyl-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, MAGNESIUM ION, Thymidylate kinase | | Authors: | Choi, J.Y, Plummer, M.S, Starr, J, Desbonnet, C.R, Soutter, H.H, Chang, J, Miller, J.R, Dillman, K, Miller, A.A, Roush, W.R. | | Deposit date: | 2011-12-02 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure guided development of novel thymidine mimetics targeting Pseudomonas aeruginosa thymidylate kinase: from hit to lead generation.
J.Med.Chem., 55, 2012
|
|
3UWO
 
 | | Structure Guided Development of Novel Thymidine Mimetics targeting Pseudomonas aeruginosa Thymidylate Kinase: from Hit to Lead Generation | | Descriptor: | 3-(1-methyl-2-oxo-2,3-dihydro-1H-imidazo[4,5-b]pyridin-6-yl)benzamide, Thymidylate kinase | | Authors: | Choi, J.Y, Plummer, M.S, Starr, J, Desbonnet, C.R, Soutter, H.H, Chang, J, Miller, J.R, Dillman, K, Miller, A.A, Roush, W.R. | | Deposit date: | 2011-12-02 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure guided development of novel thymidine mimetics targeting Pseudomonas aeruginosa thymidylate kinase: from hit to lead generation.
J.Med.Chem., 55, 2012
|
|
1YU4
 
 | | Major Tropism Determinant U1 Variant | | Descriptor: | MAGNESIUM ION, Major Tropism Determinant (Mtd-U1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YU1
 
 | | Major Tropism Determinant P3c Variant | | Descriptor: | MAGNESIUM ION, METHYL MERCURY ION, Major Tropism Determinant (Mtd-P3c) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YU0
 
 | | Major Tropism Determinant P1 Variant | | Descriptor: | CALCIUM ION, Major Tropism Determinant (Mtd-P1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YU3
 
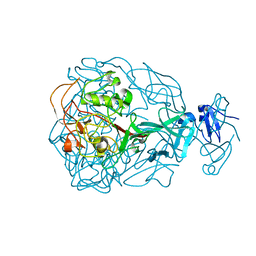 | | Major Tropism Determinant I1 Variant | | Descriptor: | MAGNESIUM ION, Major Tropism Determinant (Mtd-I1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YU2
 
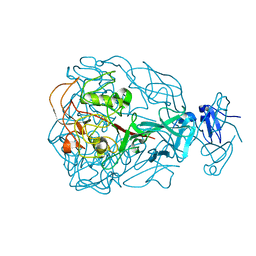 | | Major Tropism Determinant M1 Variant | | Descriptor: | MAGNESIUM ION, Major Tropism Determinant (Mtd-M1) | | Authors: | McMahon, S.A, Miller, J.L, Lawton, J.A, Ghosh, P. | | Deposit date: | 2005-02-11 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The C-type lectin fold as an evolutionary solution for massive sequence variation
Nat.Struct.Mol.Biol., 12, 2005
|
|
5IOO
 
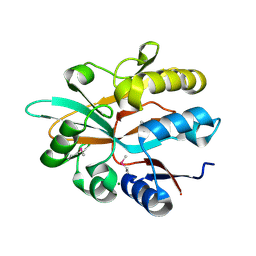 | | Accommodation of massive sequence variation in Nanoarchaeota by the C-type lectin fold | | Descriptor: | AvpA, MAGNESIUM ION | | Authors: | Handa, S, Paul, B, Miller, J, Valentine, D, Ghosh, P. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Conservation of the C-type lectin fold for accommodating massive sequence variation in archaeal diversity-generating retroelements.
Bmc Struct.Biol., 16, 2016
|
|
7LJI
 
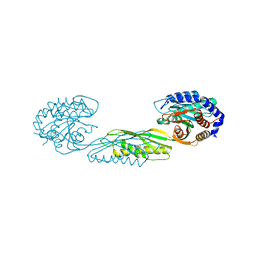 | | Structure of poly(aspartic acid) hydrolase PahZ2 with Gd+3 bound | | Descriptor: | GADOLINIUM ION, Poly(Aspartic acid) hydrolase | | Authors: | Brambley, C.A, Yared, T.J, Gonzalez, M, Jansch, A.L, Wallen, J.R, Weiland, M.H, Miller, J.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-12-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Sphingomonas sp. KT-1 PahZ2 Structure Reveals a Role for Conformational Dynamics in Peptide Bond Hydrolysis.
J.Phys.Chem.B, 125, 2021
|
|
7LJH
 
 | | Structure of poly(aspartic acid) hydrolase PahZ2 with Zn+2 bound | | Descriptor: | Poly(Aspartic acid) hydrolase, ZINC ION | | Authors: | Brambley, C.A, Yared, T.J, Gonzalez, M, Jansch, A.L, Wallen, J.R, Weiland, M.H, Miller, J.M. | | Deposit date: | 2021-01-29 | | Release date: | 2021-12-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sphingomonas sp. KT-1 PahZ2 Structure Reveals a Role for Conformational Dynamics in Peptide Bond Hydrolysis.
J.Phys.Chem.B, 125, 2021
|
|
