5PVS
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 243) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6PGT
 
 | | Protein Tyrosine Phosphatase 1B (1-301), T177A mutant, vanadate bound state | | Descriptor: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, VANADATE ION | | Authors: | Cui, D.S, Lipchock, J.M, Loria, J.P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
8E0M
 
 | |
6ULT
 
 | | BRD2-BD2 in complex with the cyclic peptide 4.2_3 | | Descriptor: | Bromodomain-containing protein 2, Cyclic peptide 4.2_3 | | Authors: | Patel, K, Walshe, J.L, Walport, L.J, Mackay, J.P. | | Deposit date: | 2019-10-08 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cyclic peptides can engage a single binding pocket through highly divergent modes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5PW8
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 259) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6M56
 
 | |
7MFW
 
 | |
6UNG
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2S)-1-(naphthalen-1-yl)-3-{[(2R)-3-oxo-2-(phenylamino)-3-{[(pyridin-3-yl)methyl]amino}propyl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2019-10-11 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
7M7E
 
 | | 6-Deoxyerythronolide B synthase (DEBS) hybrid module (M3/1) in complex with antibody fragment 1B2 | | Descriptor: | 1B2 (heavy chain), 1B2 (light chain), 6-deoxyerythronolide-B synthase EryA2, ... | | Authors: | Cogan, D.P, Zhang, K, Chiu, W, Khosla, C. | | Deposit date: | 2021-03-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Science, 374, 2021
|
|
6M5H
 
 | |
8E5T
 
 | | Yeast co-transcriptional Noc1-Noc2 RNP assembly checkpoint intermediate | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Sanghai, Z.A, Piwowarczyk, R, Vanden Broeck, A, Klinge, S. | | Deposit date: | 2022-08-22 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A co-transcriptional ribosome assembly checkpoint controls nascent large ribosomal subunit maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6PH1
 
 | | T4 lysozyme pseudo-wild type soaked in TEMPOL | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | Authors: | Cuneo, M.J, Myles, D.A, Li, L. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Making hydrogens stand out: Enhanced neutron diffraction from biological crystals using dynamic nuclear polarization
To be published
|
|
6PHA
 
 | | Protein Tyrosine Phosphatase 1B (1-301), F182A mutant, vanadate bound state | | Descriptor: | ACETATE ION, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, ... | | Authors: | Cui, D.S, Lipchock, J.M, Loria, J.P. | | Deposit date: | 2019-06-25 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
7MON
 
 | | Structure of human RIPK3-MLKL complex | | Descriptor: | Mixed lineage kinase domain-like protein, N-[4-({2-[(cyclopropanecarbonyl)amino]pyridin-4-yl}oxy)-3-fluorophenyl]-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Meng, Y, Davies, K.A, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2021-05-03 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis.
Nat Commun, 12, 2021
|
|
6M5P
 
 | | A class C beta-lactamase | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, Cha, S.S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Novel inhibition mechanism of carbapenems on the ACC-1 class C beta-lactamase.
Arch.Biochem.Biophys., 693, 2020
|
|
6ZP8
 
 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB335 | | Descriptor: | (2~{S},3~{R})-~{N}-[(5~{S},8~{S},10~{S})-5-methyl-10-oxidanyl-2,7-bis(oxidanylidene)-1,6-diazacyclododec-8-yl]-3-oxidanyl-2-(3-phenylpropanoylamino)butanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
8EBV
 
 | |
6U6O
 
 | | Crystal structure of a vaccine-elicited anti-HIV-1 rhesus macaque antibody DH846 | | Descriptor: | DH846 Fab heavy chain, DH846 Fab light chain | | Authors: | Chen, W.-H, Choe, M, Saunders, K.O, Joyce, M.G. | | Deposit date: | 2019-08-30 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structural and genetic convergence of HIV-1 neutralizing antibodies in vaccinated non-human primates.
Plos Pathog., 17, 2021
|
|
6M5X
 
 | | A fungal glyceraldehyde-3-phosphate dehydrogenase with self-resistance to inhibitor heptelidic acid | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yan, Y, Zang, X, Cooper, S.J, Lin, H, Zhou, J, Tang, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05990934 Å) | | Cite: | Biosynthesis of the fungal glyceraldehyde-3-phosphate dehydrogenase inhibitor heptelidic acid and mechanism of self-resistance
Chem Sci, 11, 2020
|
|
7MSI
 
 | |
6PHF
 
 | |
8DP3
 
 | |
7M0S
 
 | |
6M4D
 
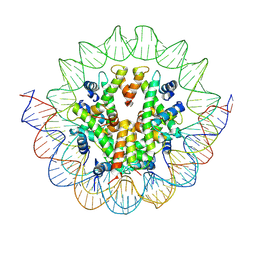 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (125-MER), Histone H2A.V, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
6ZST
 
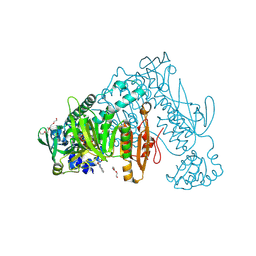 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 3-(3-methoxyquinoxalin-2-yl)propanoic acid | | Descriptor: | 3-(3-methoxyquinoxalin-2-yl)propanoic acid, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fata, F, Silvestri, I, Williams, D.L, Angelucci, F. | | Deposit date: | 2020-07-16 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the Surface of a Parasite Drug Target Thioredoxin Glutathione Reductase Using Small Molecule Fragments.
Acs Infect Dis., 7, 2021
|
|
