7LKY
 
 | |
7FW1
 
 | | Crystal Structure of human FABP4 in complex with 2-cyclopentyl-4-(4-fluorophenyl)-6-[1-(methoxymethyl)cyclopentyl]-3-methyl-5-(1H-tetrazol-5-yl)pyridine, i.e. SMILES c1(c(nc(c(c1c1ccc(cc1)F)C1=NN=NN1)C1(CCCC1)COC)C1CCCC1)C with IC50=0.0153077 microM | | Descriptor: | (5P)-2-cyclopentyl-4-(4-fluorophenyl)-6-[1-(methoxymethyl)cyclopentyl]-3-methyl-5-(1H-tetrazol-5-yl)pyridine, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kuhne, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6TG8
 
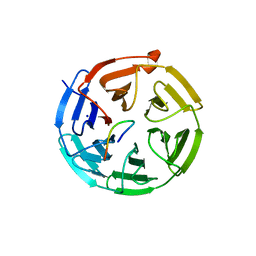 | | Crystal structure of the Kelch domain in complex with 11 amino acid peptide (model of the ETGE loop) | | Descriptor: | Kelch-like ECH-associated protein 1, SODIUM ION, VAL-ILE-ASN-PRO-GLU-THR-GLY-GLU-GLN-ILE-GLN | | Authors: | Kekez, I, Matic, S, Tomic, S, Matkovic-Calogovic, D. | | Deposit date: | 2019-11-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Binding of dipeptidyl peptidase III to the oxidative stress cell sensor Kelch-like ECH-associated protein 1 is a two-step process.
J.Biomol.Struct.Dyn., 39, 2021
|
|
5MW4
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD7 [N-(3-(((R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl)(methyl)amino)propyl)-2-(3-(2-chloro-3-(2-methylpyridin-3-yl)benzo[b]thiophen-5-yl)ureido)acetamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~2~-{[2-chloro-3-(2-methylpyridin-3-yl)-1-benzothiophen-5-yl]carbamoyl}-N-(3-{methyl[(3R)-1-(5H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]amino}propyl)glycinamide, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5IDN
 
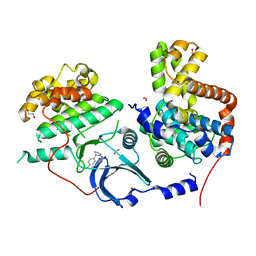 | | CDK8-CYCC IN COMPLEX WITH [(S)-2-(4-Chloro-phenyl)-pyrrolidin-1-yl]-(3-methyl-1H-pyrazolo[3,4-b]pyridin-5-yl)-methanone | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A, Czodrowski, P, Schiemann, K. | | Deposit date: | 2016-02-24 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure-Based Optimization of Potent, Selective, and Orally Bioavailable CDK8 Inhibitors Discovered by High-Throughput Screening.
J. Med. Chem., 59, 2016
|
|
8EYL
 
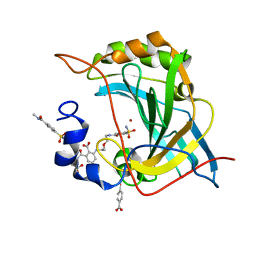 | | Human Carbonic Anhydrase II with Tert-butyl (2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethyl)carbamate | | Descriptor: | 2-[[(2~{R})-1-azanyl-5-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]carbamoyl]-6-[2-[2-[(4-sulfamoylphenyl)carbonylamino]ethoxy]ethylamino]benzoic acid, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-27 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
8K5G
 
 | | Structure of the SARS-CoV-2 BA.1 RBD with UT28-RD | | Descriptor: | Spike protein S1, UT28K-RD Fab Heavy chain, UT28K-RD Fab Light chain | | Authors: | Chen, L, Kita, S, Anraku, Y, Maenaka, K. | | Deposit date: | 2023-07-21 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Rational in silico design identifies two mutations that restore UT28K SARS-CoV-2 monoclonal antibody activity against Omicron BA.1.
Structure, 32, 2024
|
|
5IEQ
 
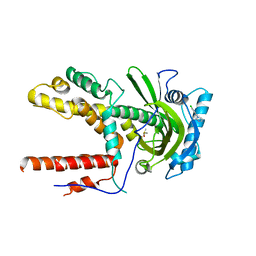 | |
3JTI
 
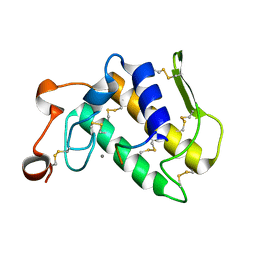 | | Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2 isoform 3, octapeptide from Amyloid beta A4 protein | | Authors: | Pandey, N, Mirza, Z, Vikram, G, Singh, N, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-09-12 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution
To be Published
|
|
6ZN5
 
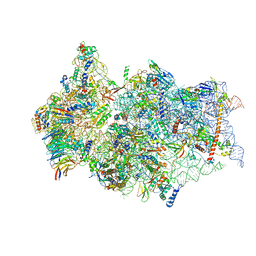 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex - state 2 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6S4G
 
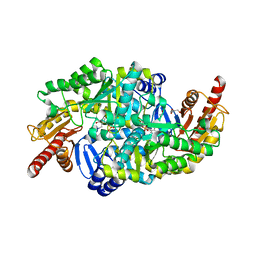 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ruggieri, F, Campillo Brocal, J.C, Humble, M.S, Walse, B, Logan, D.T, Berglund, P. | | Deposit date: | 2019-06-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Insight into the dimer dissociation process of the Chromobacterium violaceum (S)-selective amine transaminase.
Sci Rep, 9, 2019
|
|
4Z7Z
 
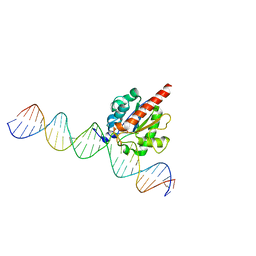 | | Structure of the enzyme-product complex resulting from TDG action on a GT mismatch in the presence of excess base | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-04-08 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
5LT7
 
 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
8EVK
 
 | | Crystal structure of Helicobacter pylori dihydroneopterin aldolase (DHNA) | | Descriptor: | 1,2-ETHANEDIOL, Dihydroneopterin aldolase, PTERINE | | Authors: | Shaw, G.X, Cherry, S, Tropea, J.E, Ji, X. | | Deposit date: | 2022-10-20 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of Helicobacter pylori dihydroneopterin aldolase suggests a fragment-based strategy for isozyme-specific inhibitor design.
Curr Res Struct Biol, 5, 2023
|
|
7T4J
 
 | | Crystal Structure of EGFR_D770_N771insNPG/V948R in complex with TAK-788 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Epidermal growth factor receptor, ... | | Authors: | Skene, R.J, Lane, W, Hu, Y. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of mobocertinib, a potent, oral inhibitor of EGFR exon 20 insertion mutations in non-small cell lung cancer.
Bioorg.Med.Chem.Lett., 80, 2022
|
|
7SZP
 
 | |
4ZKR
 
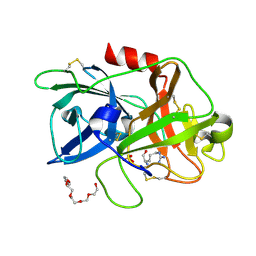 | |
6GLU
 
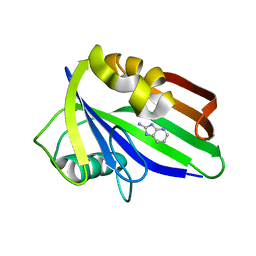 | | Crystal structure of hMTH1 D120N in complex with LW14 in the presence of acetate | | Descriptor: | 1~{H}-imidazo[4,5-b]pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Eberle, S.A, Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | hMTH1 D120N in complex with LW14
To Be Published
|
|
5I5W
 
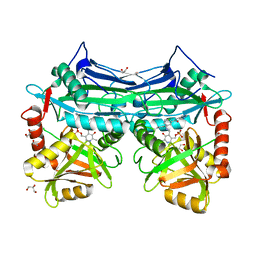 | |
6GHQ
 
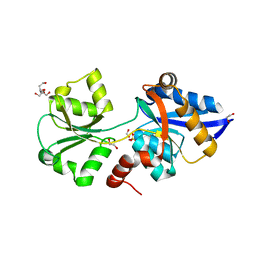 | | HtxB D206N protein variant from Pseudomonas stutzeri in a partially open conformation to 1.53 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
8RLJ
 
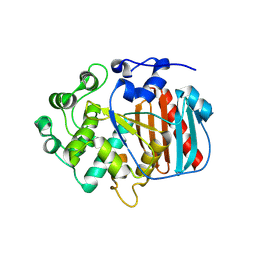 | | Structure of the apo form of PIB-1 in an Orthorombic space group | | Descriptor: | Class C beta-lactamase-related serine hydrolase | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A new type of Class C beta-lactamases defined by PIB-1. A metal-dependent carbapenem-hydrolyzing beta-lactamase, from Pseudomonas aeruginosa: Structural and functional analysis.
Int.J.Biol.Macromol., 277, 2024
|
|
7O2C
 
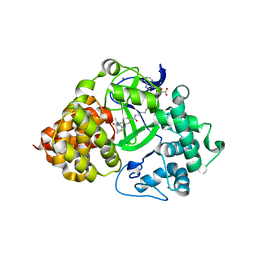 | | X-RAY STRUCTURE OF SMYD3 IN COMPLEX WITH the benzodiazepine-based probe BAY-6035 | | Descriptor: | (2S)-1-[[(1R,5S)-3-azabicyclo[3.1.0]hexan-3-yl]carbonyl]-N-(2-cyclopropylethyl)-2-methyl-4-oxidanylidene-3,5-dihydro-2H-1,5-benzodiazepine-7-carboxamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Steuber, H. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of the SMYD3 Inhibitor BAY-6035 Using Thermal Shift Assay (TSA)-Based High-Throughput Screening.
Slas Discov, 26, 2021
|
|
8RLL
 
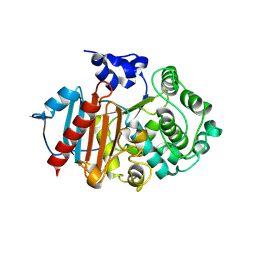 | | Structure of the apo form of PIB-1 in an Orthorombic space group | | Descriptor: | Class C beta-lactamase-related serine hydrolase | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A new type of Class C beta-lactamases defined by PIB-1. A metal-dependent carbapenem-hydrolyzing beta-lactamase, from Pseudomonas aeruginosa: Structural and functional analysis.
Int.J.Biol.Macromol., 277, 2024
|
|
8YGD
 
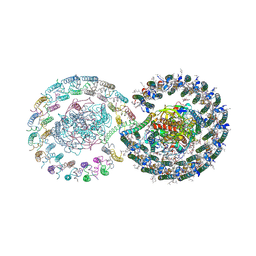 | | Rhodobacter blasticus RC-LH1 dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-deoxy-D-xylulose-5-phosphate synthase, Antenna pigment protein alpha chain, ... | | Authors: | Liu, L.N, Zhang, Y.Z, Wang, P, Christianson, B.M, Ugurlar, D. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Architectures of photosynthetic RC-LH1 supercomplexes from Rhodobacter blasticus.
Sci Adv, 10, 2024
|
|
8YGL
 
 | | Rhodobacter blasticus RC-LH1 monomer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-deoxy-D-xylulose-5-phosphate synthase, Antenna pigment protein alpha chain, ... | | Authors: | Liu, L.N, Zhang, Y.Z, Wang, P, Christianson, B.M, Ugurlar, D. | | Deposit date: | 2024-02-26 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Architectures of photosynthetic RC-LH1 supercomplexes from Rhodobacter blasticus.
Sci Adv, 10, 2024
|
|
