7R0W
 
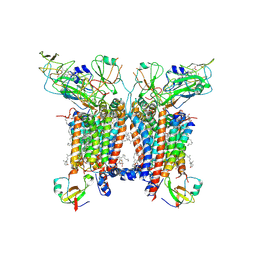 | | 2.8 Angstrom cryo-EM structure of the dimeric cytochrome b6f-PetP complex from Synechocystis sp. PCC 6803 with natively bound lipids and plastoquinone molecules | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S,8E)-1-{[(2S)-1-hydroxy-3-{[(1S)-1-hydroxypentadecyl]oxy}propan-2-yl]oxy}heptadec-8-en-1-ol, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, ... | | Authors: | Farmer, D.F, Proctor, M.S, Malone, L.A, Swainsbury, D.P.K, Hawkings, F.R, Hitchcock, A, Johnson, M.P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of the Synechocystis sp. PCC 6803 cytochrome b6f complex with and without the regulatory PetP subunit.
Biochem.J., 479, 2022
|
|
8ASJ
 
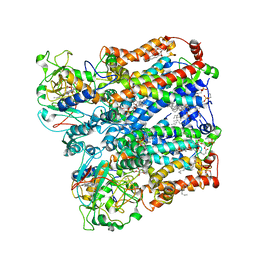 | | Four subunit cytochrome b-c1 complex from Rhodobacter sphaeroides in native nanodiscs - focussed refinement in the b-c conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Cytochrome b, Cytochrome b-c1 subunit IV, ... | | Authors: | Swainsbury, D.J.K, Hawkings, F.R, Martin, E.C, Musial, S, Salisbury, J.H, Jackson, P.J, Farmer, D.A, Johnson, M.P, Siebert, C.A, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structure of the four-subunit Rhodobacter sphaeroides cytochrome bc 1 complex in styrene maleic acid nanodiscs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6RQF
 
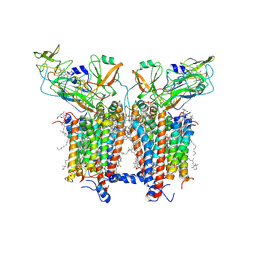 | | 3.6 Angstrom cryo-EM structure of the dimeric cytochrome b6f complex from Spinacia oleracea with natively bound thylakoid lipids and plastoquinone molecules | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Malone, L.A, Qian, P, Mayneord, G.E, Hitchcock, A, Farmer, D, Thompson, R, Swainsbury, D.J.K, Ranson, N, Hunter, C.N, Johnson, M.P. | | Deposit date: | 2019-05-15 | | Release date: | 2019-11-13 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structure of the spinach cytochrome b6f complex at 3.6 angstrom resolution.
Nature, 575, 2019
|
|
7ZXY
 
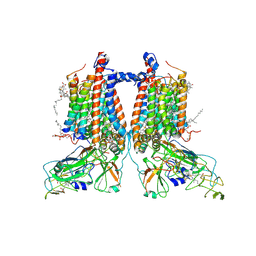 | | 3.15 Angstrom cryo-EM structure of the dimeric cytochrome b6f complex from Synechocystis sp. PCC 6803 with natively bound plastoquinone and lipid molecules. | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, CHLOROPHYLL A, Cytochrome B6, ... | | Authors: | Malone, L.A, Procter, M.S, Farmer, D.F, Swainsbury, D.J.K, Hawkings, F.R, Pastorelli, F, Emrich-Mills, T.Z, Siebert, A, Hunter, C.N, Hitchcock, A, Johnson, M.P. | | Deposit date: | 2022-05-23 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structures of the Synechocystis sp. PCC 6803 cytochrome b6f complex with and without the regulatory PetP subunit.
Biochem.J., 479, 2022
|
|
6Z5S
 
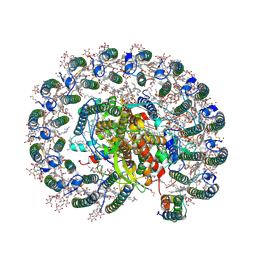 | | RC-LH1(14)-W complex from Rhodopseudomonas palustris | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, (6~{R},10~{S},14~{R},19~{R},23~{S},24~{E},27~{S},28~{E})-2,6,10,14,19,23,27,31-octamethyldotriaconta-24,28-dien-2-ol, ... | | Authors: | Swainsbury, D.J.K, Qian, P, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2020-05-27 | | Release date: | 2021-01-13 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structures of Rhodopseudomonas palustris RC-LH1 complexes with open or closed quinone channels.
Sci Adv, 7, 2021
|
|
6Z5R
 
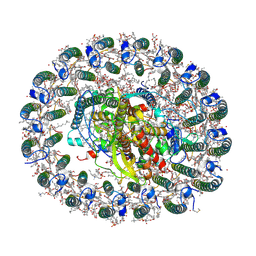 | | RC-LH1(16) complex from Rhodopseudomonas palustris | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, (6~{R},10~{S},14~{R},19~{R},23~{S},24~{E},27~{S},28~{E})-2,6,10,14,19,23,27,31-octamethyldotriaconta-24,28-dien-2-ol, ... | | Authors: | Swainsbury, D.J.K, Qian, P, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2020-05-27 | | Release date: | 2021-01-13 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of Rhodopseudomonas palustris RC-LH1 complexes with open or closed quinone channels.
Sci Adv, 7, 2021
|
|
8ASI
 
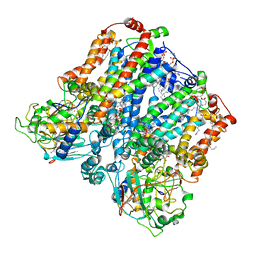 | | Four subunit cytochrome b-c1 complex from Rhodobacter sphaeroides in native nanodiscs - consensus refinement in the b-b conformation | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Cytochrome b, Cytochrome b-c1 subunit IV, ... | | Authors: | Swainsbury, D.J.K, Hawkings, F.R, Martin, E.C, Musial, S, Salisbury, J.H, Jackson, P.J, Farmer, D.A, Johnson, M.P, Siebert, C.A, Hitchcock, A, Hunter, C.N. | | Deposit date: | 2022-08-19 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the four-subunit Rhodobacter sphaeroides cytochrome bc 1 complex in styrene maleic acid nanodiscs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5O37
 
 | |
5O2J
 
 | |
5O2K
 
 | |
5LQ5
 
 | | 1.46 A resolution structure of PhnD1 from Prochlorococcus marinus (MIT 9301) in complex with phosphite | | Descriptor: | PHOSPHITE ION, Putative phosphonate binding protein for ABC transporter | | Authors: | Bisson, C, Adams, N.B.P, Polyviou, D, Bibby, T.S, Hunter, C.N, Hitchcock, A. | | Deposit date: | 2016-08-16 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The molecular basis of phosphite and hypophosphite recognition by ABC-transporters.
Nat Commun, 8, 2017
|
|
5LQ8
 
 | | 1.52 A resolution structure of PhnD1 from Prochlorococcus marinus (MIT 9301) in complex with methylphosphonate | | Descriptor: | METHYLPHOSPHONIC ACID ESTER GROUP, Putative phosphonate binding protein for ABC transporter | | Authors: | Bisson, C, Adams, N.B.P, Polyviou, D, Bibby, T.S, Hunter, C.N, Hitchcock, A. | | Deposit date: | 2016-08-16 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The molecular basis of phosphite and hypophosphite recognition by ABC-transporters.
Nat Commun, 8, 2017
|
|
5LQ1
 
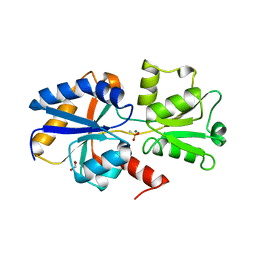 | | 1.41 A resolution structure of PtxB from Trichodesmium erythraeum IMS101 in complex with methylphosphonate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, METHYLPHOSPHONIC ACID ESTER GROUP, ... | | Authors: | Bisson, C, Adams, N.B.P, Polyviou, D, Bibby, T.S, Hunter, C.N, Hitchcock, A. | | Deposit date: | 2016-08-15 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The molecular basis of phosphite and hypophosphite recognition by ABC-transporters.
Nat Commun, 8, 2017
|
|
5LV1
 
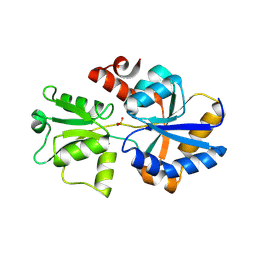 | | 2.12 A resolution structure of PtxB from Prochlorococcus marinus (MIT 9301) in complex with phosphite | | Descriptor: | PtxB, oxidanylphosphinate | | Authors: | Bisson, C, Adams, N.B.P, Polyviou, D, Bibby, T.S, Hunter, C.N, Hitchcock, A. | | Deposit date: | 2016-09-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The molecular basis of phosphite and hypophosphite recognition by ABC-transporters.
Nat Commun, 8, 2017
|
|
5JVB
 
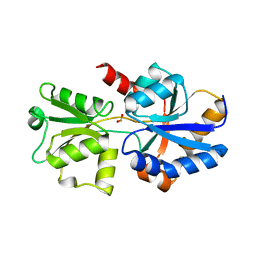 | | 1.95A resolution structure of PtxB from Trichodesmium erythraeum IMS101 in complex with phosphite | | Descriptor: | PHOSPHONATE, Phosphonate ABC transporter, periplasmic phosphonate-binding protein | | Authors: | Bisson, C, Adams, N.B.P, Polyviou, D, Bibby, T.S, Hunter, C.N, Hitchcock, A. | | Deposit date: | 2016-05-11 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The molecular basis of phosphite and hypophosphite recognition by ABC-transporters.
Nat Commun, 8, 2017
|
|
5ME4
 
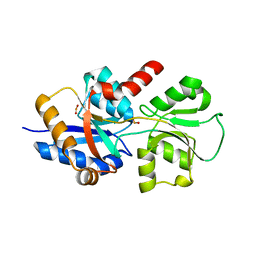 | |
7NRR
 
 | | The structure of the SBP TarP_Csal in complex with caffeate | | Descriptor: | CAFFEIC ACID, MAGNESIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-04 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NQG
 
 | | The structure of the SBP TarP_Rhp in complex with 4-hydroxyphenylacetate | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENYLACETATE, PHOSPHATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NTE
 
 | | The structure of an open conformation of the SBP TarP_Csal | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NSW
 
 | | The structure of the SBP TarP_Csal in complex with coumarate | | Descriptor: | 1,2-ETHANEDIOL, 4'-HYDROXYCINNAMIC ACID, MAGNESIUM ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NRA
 
 | | The structure of the SBP TarP_Sse in complex with cinnamate | | Descriptor: | HYDROCINNAMIC ACID, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-03 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NTD
 
 | | The structure of the SBP TarP_Csal in complex with ferulate | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7NR2
 
 | | The structure of the SBP TarP_Sse in complex with coumarate | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, SULFATE ION, TRAP dicarboxylate transporter, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-10-06 | | Last modified: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
6EMN
 
 | | HtxB from Pseudomonas stutzeri in complex with phosphite to 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable phosphite transport system-binding protein HtxB, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2017-10-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
6GHQ
 
 | | HtxB D206N protein variant from Pseudomonas stutzeri in a partially open conformation to 1.53 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
