3UV3
 
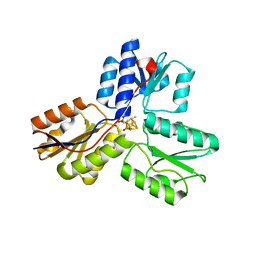 | | Ec_IspH in complex with but-2-ynyl diphosphate (1086) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, but-2-yn-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
4BKU
 
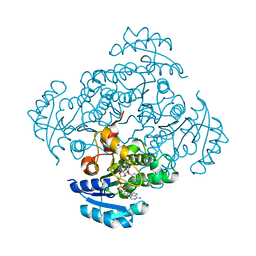 | | Enoyl-ACP reductase FabI from Burkholderia pseudomallei with cofactor NADH and inhibitor PT155 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 5-(4-amino-2-methylphenoxy)-2-hexyl-4-hydroxy-1-methylpyridinium, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH] | | Authors: | Hirschbeck, M.W, Liu, N, Neckles, C, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-04-29 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Rational Design of Broad Spectrum Antibacterial Activity Based on a Clinically Relevant Enoyl-Acyl Carrier Protein (Acp) Reductase Inhibitor.
J.Biol.Chem., 289, 2014
|
|
8BQS
 
 | | Cryo-EM structure of the I-II-III2-IV2 respiratory supercomplex from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Baradaran, R, Amunts, A. | | Deposit date: | 2022-11-21 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
4BIO
 
 | |
4CCK
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in complex with Mn(II) and N-oxalylglycine (NOG) | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCJ
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in apo form | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66 | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
7GIX
 
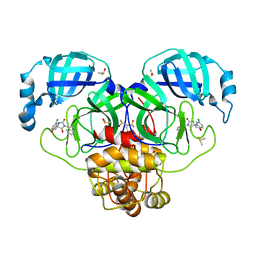 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with VLA-UNK-cf7facf1-1 (Mpro-P0143) | | Descriptor: | (1'M,4S)-6-chloro-1'-(isoquinolin-4-yl)-2,3-dihydrospiro[[1]benzopyran-4,4'-imidazolidine]-2',5'-dione, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
2RBE
 
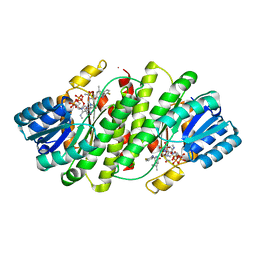 | | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors | | Descriptor: | (5R)-2-[(2-fluorophenyl)amino]-5-(1-methylethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Jordan, S.R, Li, V. | | Deposit date: | 2007-09-18 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2GQX
 
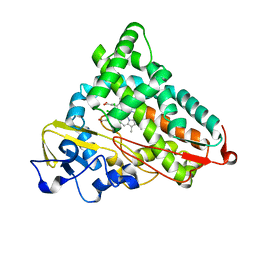 | | Crystal structure of cytochrome p450cam mutant (f87w/y96f/l244a/v247l/c334a) with pentachlorobenzene | | Descriptor: | 1,2,3,4,5-PENTACHLOROBENZENE, Cytochrome P450-cam, POTASSIUM ION, ... | | Authors: | Rao, Z, Wong, L.L, Xu, F, Bell, S.G. | | Deposit date: | 2006-04-22 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity correlations in pentachlorobenzene oxidation by engineered cytochrome P450cam
Protein Eng.Des.Sel., 20, 2007
|
|
6C49
 
 | |
5GLY
 
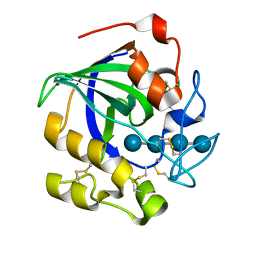 | | Crystal structure of a glycoside hydrolase in complex with cellotetrose from Thielavia terrestris NRRL 8126 | | Descriptor: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
3ESS
 
 | |
1ZKP
 
 | | 1.5A Resolution Crystal Structure of a Metallo Beta Lactamase Family Protein, the ELAC Homolgue of Bacillus anthracis, a Putative Ribonuclease | | Descriptor: | CHLORIDE ION, SODIUM ION, ZINC ION, ... | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | 1.5A Resolution Crystal Structure of a Metallo Beta Lactamase Family
Protein, the ELAC Homolgue of Bacillus anthracis, a Putative Ribonuclease
To be Published
|
|
7Z7T
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z83
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZC5
 
 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
1ZOX
 
 | |
1DSE
 
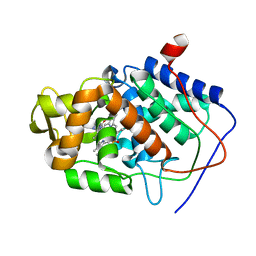 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX, WITH PHOSPHATE BOUND, PH 6, 100K | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
7Z84
 
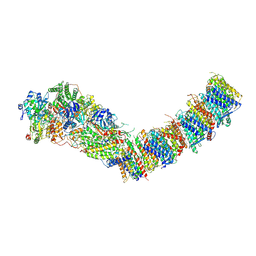 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
3URK
 
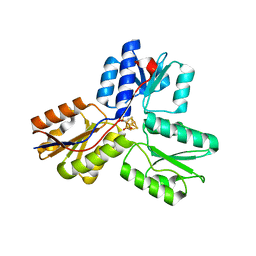 | | IspH in complex with propynyl diphosphate (1061) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, prop-2-yn-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-22 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
7Z80
 
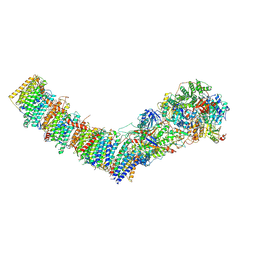 | | Complex I from E. coli, DDM/LMNG-purified, under Turnover at pH 8, Closed state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z3E
 
 | | XFEL structure of Class Ib ribonucleotide reductase dimanganese(II) NrdF in complex with hydroquinone NrdI from Bacillus cereus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, MANGANESE (II) ION, Protein NrdI, ... | | Authors: | John, J, Lebrette, H, Aurelius, O, Hogbom, M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-controlled reorganization and flavin strain within the ribonucleotide reductase R2b-NrdI complex monitored by serial femtosecond crystallography.
Elife, 11, 2022
|
|
7Z7V
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7Z7R
 
 | | Complex I from E. coli, LMNG-purified, Apo, Open-ready state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7ZCI
 
 | | Complex I from E. coli, LMNG-purified, under Turnover at pH 6, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2022-03-28 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
