5TCL
 
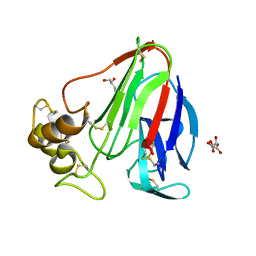 | | Thaumatin from 4.96 A wavelength data collection | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Aurelius, O, Duman, R, El Omari, K, Mykhaylyk, V, Wagner, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Long-wavelength macromolecular crystallography - First successful native SAD experiment close to the sulfur edge.
Nucl Instrum Methods Phys Res B, 411, 2017
|
|
7BET
 
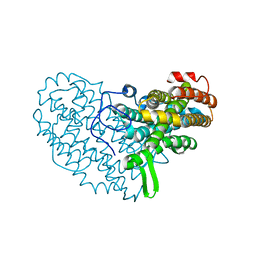 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Hogbom, M, Wang, M, Pedrini, B. | | Deposit date: | 2020-12-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
7AI9
 
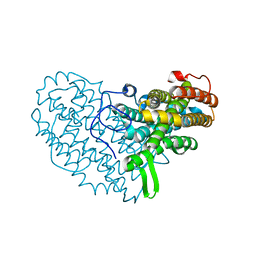 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by rotation serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Hogbom, M, Wang, M, Marsh, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AI8
 
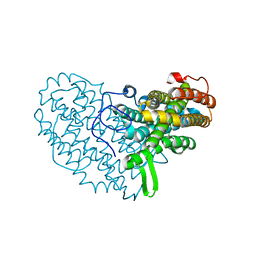 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by still serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Hogbom, M, Wang, M, Marsh, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4COJ
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima in complex with dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4COI
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with glycerol in the active site | | Descriptor: | ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, GLYCEROL, ZINC ION | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-28 | | Release date: | 2015-01-14 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4COM
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with MES in the active site | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4COL
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with dATP bound in the specificity site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, MAGNESIUM ION, ... | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
4CON
 
 | | Crystal structure of the anaerobic ribonucleotide reductase from Thermotoga maritima with citrate in the active site | | Descriptor: | ANAEROBIC RIBONUCLEOSIDE-TRIPHOSPHATE REDUCTASE, CITRIC ACID | | Authors: | Aurelius, O, Johansson, R, Bagenholm, V, Beck, T, Balhuizen, A, Lundin, D, Sjoberg, B.M, Mulliez, E, Logan, D.T. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Crystal Structure of Thermotoga Maritima Class III Ribonucleotide Reductase Lacks a Radical Cysteine Pre-Positioned in the Active Site.
Plos One, 10, 2015
|
|
5IUQ
 
 | | Galectin-3c in complex with Bisamido-thiogalactoside derivative 4 | | Descriptor: | 3-deoxy-3-[(2,3,5,6-tetrafluoro-4-methoxybenzene-1-carbonyl)amino]-beta-D-galactopyranosyl 3-deoxy-3-[(2,3,5,6-tetrafluoro-4-methoxybenzene-1-carbonyl)amino]-1-thio-beta-D-galactopyranoside, Galectin-3 | | Authors: | Noresson, A.-L, Aurelius, O, Oberg, C.T, Engstrom, O, Sundin, A.P, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2016-03-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.121 Å) | | Cite: | Controlling protein:ligand complex conformation through tuning of arginine-arene interactions: Synthetic and structural studies with 3-benzamido-2-sulfo-galactosides as galectin-3 ligands
To Be Published
|
|
4ZXO
 
 | | The structure of a GH26 beta-mannanase from Bacteroides ovatus, BoMan26A. | | Descriptor: | Glycosyl hydrolase family 26, PHOSPHATE ION, POTASSIUM ION | | Authors: | Bagenholm, V, Aurelius, O, Logan, D.T, Bouraoui, H, Stalbrand, H. | | Deposit date: | 2015-05-20 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Galactomannan Catabolism Conferred by a Polysaccharide Utilization Locus of Bacteroides ovatus: ENZYME SYNERGY AND CRYSTAL STRUCTURE OF A beta-MANNANASE.
J. Biol. Chem., 292, 2017
|
|
5EHB
 
 | | A de novo designed hexameric coiled-coil peptide with iodotyrosine | | Descriptor: | pHiosYI | | Authors: | Lizatovic, R, Aurelius, O, Stenstrom, O, Drakenberg, T, Akke, M, Logan, D.T, Andre, I. | | Deposit date: | 2015-10-28 | | Release date: | 2016-06-15 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | A De Novo Designed Coiled-Coil Peptide with a Reversible pH-Induced Oligomerization Switch.
Structure, 24, 2016
|
|
8PUR
 
 | | DexyHemoglobin structure from serial synchrotron crystallography with fixed target | | Descriptor: | Hemoglobin subunit beta, Hemoglobin, alpha 1, ... | | Authors: | Bjelcic, M, Sigfridsson Clauss, K, Aurelius, O, Milas, M, Nan, J, Ursby, T. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Anaerobic fixed-target serial crystallography using sandwiched silicon nitride membranes.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8PUQ
 
 | | MetHemoglobin structure from serial synchrotron crystallography with fixed target | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bjelcic, M, Sigfridsson Clauss, K, Aurelius, O, Milas, M, Nan, J, Ursby, T. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Anaerobic fixed-target serial crystallography using sandwiched silicon nitride membranes.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z3E
 
 | | XFEL structure of Class Ib ribonucleotide reductase dimanganese(II) NrdF in complex with hydroquinone NrdI from Bacillus cereus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, MANGANESE (II) ION, Protein NrdI, ... | | Authors: | John, J, Lebrette, H, Aurelius, O, Hogbom, M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-controlled reorganization and flavin strain within the ribonucleotide reductase R2b-NrdI complex monitored by serial femtosecond crystallography.
Elife, 11, 2022
|
|
7Z3D
 
 | | XFEL structure of Class Ib ribonucleotide reductase dimanganese(II) NrdF in complex with oxidized NrdI from Bacillus cereus | | Descriptor: | FLAVIN MONONUCLEOTIDE, MANGANESE (II) ION, Protein NrdI, ... | | Authors: | John, J, Lebrette, H, Aurelius, O, Hogbom, M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Redox-controlled reorganization and flavin strain within the ribonucleotide reductase R2b-NrdI complex monitored by serial femtosecond crystallography.
Elife, 11, 2022
|
|
8BBR
 
 | | Determination of the structure of active tyrosinase from bacterium Verrucomicrobium spinosum | | Descriptor: | COPPER (II) ION, Core tyrosinase, SULFATE ION | | Authors: | Fekry, M, Dave, K, Badgujar, D, Aurelius, O, Hamnevik, E, Dobritzsch, D, Danielson, H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Crystal Structure of Tyrosinase from Verrucomicrobium spinosum Reveals It to Be an Atypical Bacterial Tyrosinase.
Biomolecules, 13, 2023
|
|
8BBQ
 
 | | Determination of the structure of active tyrosinase from bacterium Verrucomicrobium spinosum | | Descriptor: | COPPER (II) ION, Core tyrosinase, GLYCEROL, ... | | Authors: | Fekry, M, Dave, K, Badgujar, D, Aurelius, O, Hamnevik, E, Dobritzsch, D, Danielson, H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The Crystal Structure of Tyrosinase from Verrucomicrobium spinosum Reveals It to Be an Atypical Bacterial Tyrosinase.
Biomolecules, 13, 2023
|
|
6Y2N
 
 | | Crystal structure of ribonucleotide reductase R2 subunit solved by serial synchrotron crystallography | | Descriptor: | FE (III) ION, MANGANESE (III) ION, Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Shilova, A, Lebrette, H, Aurelius, O, Hogbom, M, Mueller, U. | | Deposit date: | 2020-02-17 | | Release date: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
