8HFO
 
 | | Crystal Structure of Mycobacterium smegmatis MshC in Complex with Compound 7d | | Descriptor: | CALCIUM ION, L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, N-[(3M)-3-(thiophen-2-yl)benzene-1-sulfonyl]-L-cysteinamide, ... | | Authors: | Pang, L, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Basis of Cysteine Ligase MshC Inhibition by Cysteinyl-Sulfonamides.
Int J Mol Sci, 23, 2022
|
|
6S4T
 
 | | LXRbeta ligand binding domain in comlpex with small molecule inhibitors | | Descriptor: | 2-[4-[[3-[3-(phenylmethyl)-8-(trifluoromethyl)quinolin-4-yl]phenoxy]methyl]phenyl]ethanoic acid, Oxysterols receptor LXR-beta | | Authors: | Sandmark, J, Jansson, A. | | Deposit date: | 2019-06-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands.
Commun Biol, 2, 2019
|
|
6S5K
 
 | | LXRbeta ligand binding domain in complex with small molecule inhibitors | | Descriptor: | 3-(4-phenylbutylamino)-1,4-bis(phenylmethyl)pyrrole-2,5-dione, Oxysterols receptor LXR-beta | | Authors: | Petersen, J. | | Deposit date: | 2019-07-01 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands.
Commun Biol, 2, 2019
|
|
6S4U
 
 | | LXRbeta ligand binding domain in comlpex with small molecule inhibitors | | Descriptor: | 6-[4-[[3-oxidanyl-1,1-bis(oxidanylidene)-5-phenyl-2-propan-2-yl-3~{H}-1,2-thiazol-4-yl]amino]butyl]pyridine-2-sulfonamide, Oxysterols receptor LXR-beta | | Authors: | Sandmark, J, Jansson, A. | | Deposit date: | 2019-06-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands.
Commun Biol, 2, 2019
|
|
8ZFQ
 
 | |
8ZFP
 
 | |
8ZFO
 
 | |
8ZFR
 
 | |
8ZFT
 
 | |
8ZFN
 
 | |
8ZFS
 
 | |
6S4N
 
 | | LXRbeta ligand binding domain in comlpex with small molecule inhibitors | | Descriptor: | 2-[5-chloranyl-6-[4-[[1,1,3-tris(oxidanylidene)-5-phenyl-2-propan-2-yl-1,2-thiazol-4-yl]amino]piperidin-1-yl]pyridin-3-yl]ethanoic acid, Oxysterols receptor LXR-beta, SULFATE ION | | Authors: | Sandmark, J, Jansson, A. | | Deposit date: | 2019-06-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis identifies an escape route from the adverse lipogenic effects of liver X receptor ligands.
Commun Biol, 2, 2019
|
|
2TYS
 
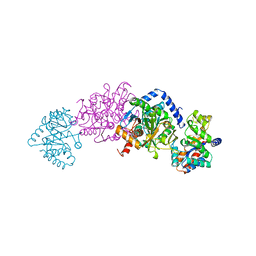 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-L-TRYPTOPHANE | | Authors: | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1997-01-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
1D2R
 
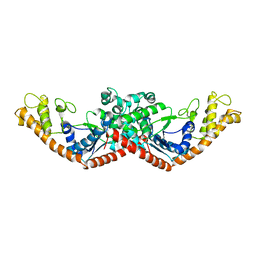 | |
1N4H
 
 | | Characterization of ligands for the orphan nuclear receptor RORbeta | | Descriptor: | Nuclear Receptor ROR-beta, RETINOIC ACID, Steroid Receptor Coactivator-1 | | Authors: | Stehlin-Gaon, C, Willmann, D, Sanglier, S, Van Dorsselaer, A, Renaud, J.-P, Moras, D, Schuele, R. | | Deposit date: | 2002-10-31 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | All-trans retinoic acid is a ligand for the orphan nuclear receptor RORbeta
Nat.Struct.Biol., 10, 2003
|
|
1N0T
 
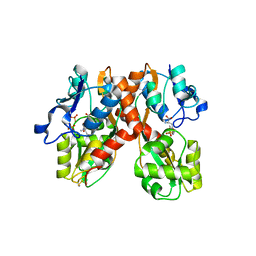 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with the antagonist (S)-ATPO at 2.1 A resolution. | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Greenwood, J.R, Liljefors, T, Lunn, M.-L, Egebjerg, J, Larsen, I.K, Gouaux, E, Kastrup, J.S. | | Deposit date: | 2002-10-15 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive antagonism of AMPA receptors by ligands of
different classes: crystal structure of ATPO bound to the
GluR2 ligand-binding core, in comparison with DNQX.
J.Med.Chem., 46, 2003
|
|
1AEK
 
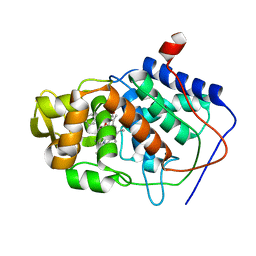 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (INDOLINE) | | Descriptor: | CYTOCHROME C PEROXIDASE, INDOLINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
1AEE
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (ANILINE) | | Descriptor: | ANILINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
1AEN
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (2-AMINO-5-METHYLTHIAZOLE) | | Descriptor: | 2-AMINO-5-METHYLTHIAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Bunte, S.W, Rosenfeld, R, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
1AEF
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (3-AMINOPYRIDINE) | | Descriptor: | 3-AMINOPYRIDINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
1AEG
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (4-AMINOPYRIDINE) | | Descriptor: | 4-AMINOPYRIDINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
1AEM
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (IMIDAZO[1,2-A]PYRIDINE) | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZO[1,2-A]PYRIDINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
4ZN7
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with Diethylstilbestrol | | Descriptor: | DIETHYLSTILBESTROL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.934 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
1GR2
 
 | | STRUCTURE OF A GLUTAMATE RECEPTOR LIGAND BINDING CORE (GLUR2) COMPLEXED WITH KAINATE | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, PROTEIN (GLUTAMATE RECEPTOR 2) | | Authors: | Armstrong, N, Sun, Y, Chen, G.Q, Gouaux, E. | | Deposit date: | 1998-09-17 | | Release date: | 1998-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a glutamate-receptor ligand-binding core in complex with kainate.
Nature, 395, 1998
|
|
1AEH
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (2-AMINO-4-METHYLTHIAZOLE) | | Descriptor: | 2-AMINO-4-METHYLTHIAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
