1E9N
 
 | | A second divalent metal ion in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1, and its implications for the catalytic mechanism | | Descriptor: | DNA-(APURINIC OR APYRIMIDINIC SITE) LYASE, LEAD (II) ION | | Authors: | Beernink, P.T, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-10-24 | | Release date: | 2001-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism
J.Mol.Biol., 307, 2001
|
|
1EV1
 
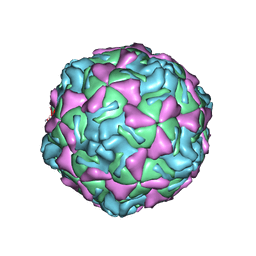 | | ECHOVIRUS 1 | | Descriptor: | ECHOVIRUS 1, MYRISTIC ACID, PALMITIC ACID | | Authors: | Wien, M.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-12-02 | | Release date: | 1999-01-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure determination of echovirus 1.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1DYH
 
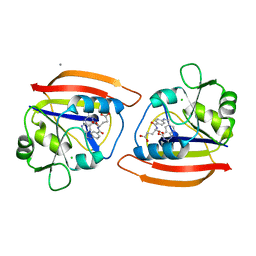 | | ISOMORPHOUS CRYSTAL STRUCTURES OF ESCHERICHIA COLI DIHYDROFOLATE REDUCTASE COMPLEXED WITH FOLATE, 5-DEAZAFOLATE AND 5,10-DIDEAZATETRAHYDROFOLATE: MECHANISTIC IMPLICATIONS | | Descriptor: | 5-DEAZAFOLIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reyes, V.M, Kraut, J. | | Deposit date: | 1994-10-26 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Isomorphous crystal structures of Escherichia coli dihydrofolate reductase complexed with folate, 5-deazafolate, and 5,10-dideazatetrahydrofolate: mechanistic implications.
Biochemistry, 34, 1995
|
|
1DYI
 
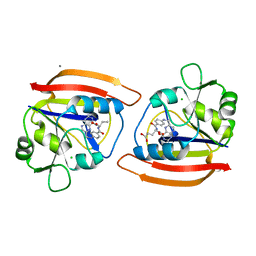 | | ISOMORPHOUS CRYSTAL STRUCTURES OF ESCHERICHIA COLI DIHYDROFOLATE REDUCTASE COMPLEXED WITH FOLATE, 5-DEAZAFOLATE AND 5,10-DIDEAZATETRAHYDROFOLATE: MECHANISTIC IMPLICATIONS | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Reyes, V.M, Kraut, J. | | Deposit date: | 1994-10-26 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Isomorphous crystal structures of Escherichia coli dihydrofolate reductase complexed with folate, 5-deazafolate, and 5,10-dideazatetrahydrofolate: mechanistic implications.
Biochemistry, 34, 1995
|
|
4LZ2
 
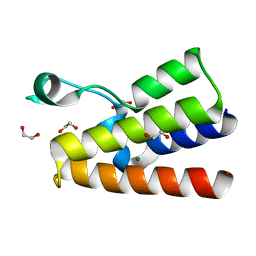 | | Crystal structure of the bromodomain of human BAZ2A | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2A, MAGNESIUM ION | | Authors: | Tallant, C, Nunez-Alonso, G, Picaud, S, Filippakopoulos, P, Krojer, T, Bradley, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-31 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
7EPT
 
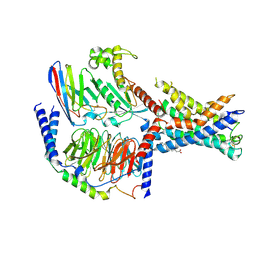 | | Structural basis for the tethered peptide activation of adhesion GPCRs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ping, Y.-Q, Xiao, P, Yang, F, Zhao, R.-J, Guo, S.-C, Yan, X, Wu, X, Liebscher, I, Xu, H.E, Sun, J.-P. | | Deposit date: | 2021-04-27 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
7F92
 
 | | Structure of connexin43/Cx43/GJA1 gap junction intercellular channel in LMNG/CHS detergents at pH ~8.0 | | Descriptor: | Gap junction alpha-1 protein, TETRADECANE | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2021-07-03 | | Release date: | 2022-07-06 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
3SZ2
 
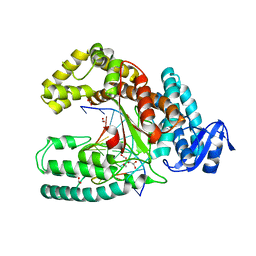 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dG as templating nucleobase | | Descriptor: | (5'-D(*AP*AP*AP*GP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3'), DNA polymerase I, ... | | Authors: | Betz, K, Marx, A, Diederichs, K. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
1CSX
 
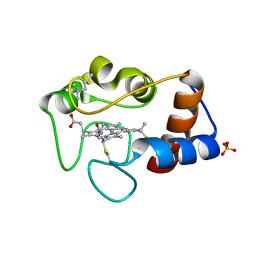 | |
1CSV
 
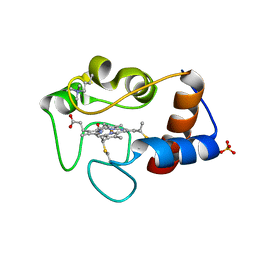 | |
1CSW
 
 | |
1CSU
 
 | |
1CLH
 
 | |
3SV3
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with the artificial base pair dNaM-d5SICSTP | | Descriptor: | (5'-D(*AP*AP*AP*(BMN)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), 2-{2-deoxy-5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-6-methylisoquinoline-1(2H)-thione, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
1G7F
 
 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH PNU177496 | | Descriptor: | 2-{4-[(2S)-2-[({[(1S)-1-CARBOXY-2-PHENYLETHYL]AMINO}CARBONYL)AMINO]-3-OXO-3-(PENTYLAMINO)PROPYL]PHENOXY}MALONIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Bleasdale, J.E, Ogg, D, Larsen, S.D. | | Deposit date: | 2000-11-10 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule peptidomimetics containing a novel phosphotyrosine bioisostere inhibit protein tyrosine phosphatase 1B and augment insulin action.
Biochemistry, 40, 2001
|
|
3SV4
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dT as templating nucleobase | | Descriptor: | (5'-D(*AP*AP*AP*TP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA polymerase I, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
4KTL
 
 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with 4,4'-(3-((4-hydroxyphenyl)amino)-1H-pyrazole-4,5-diyl)diphenol | | Descriptor: | 4,4'-{3-[(4-hydroxyphenyl)amino]-1H-pyrazole-4,5-diyl}diphenol, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A. | | Deposit date: | 2013-05-20 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Overcoming the Limitations of Fragment Merging: Rescuing a Strained Merged Fragment Series Targeting Mycobacterium tuberculosis CYP121.
Chemmedchem, 8, 2013
|
|
4KTJ
 
 | |
3SYZ
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with dNaM as templating nucleobase | | Descriptor: | (5'-D(*AP*AP*AP*(BMN)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA polymerase I, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry.
Nat.Chem.Biol., 8, 2012
|
|
4N7E
 
 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with AF38469 | | Descriptor: | 2-[(6-methylpyridin-2-yl)carbamoyl]-5-(trifluoromethyl)benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2013-10-15 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The identification of AF38469: An orally bioavailable inhibitor of the VPS10P family sorting receptor Sortilin.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4KTK
 
 | |
1D7O
 
 | | CRYSTAL STRUCTURE OF BRASSICA NAPUS ENOYL ACYL CARRIER PROTEIN REDUCTASE COMPLEXED WITH NAD AND TRICLOSAN | | Descriptor: | ENOYL-[ACYL-CARRIER PROTEIN] REDUCTASE (NADH) PRECURSOR, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Roujeinikova, A, Levy, C, Rowsell, S, Sedelnikova, S, Baker, P.J, Minshull, C.A, Mistry, A, Colls, J.G, Camble, R, Stuitje, A.R, Slabas, A.R, Rafferty, J.B, Pauptit, R.A, Viner, R, Rice, D.W. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis of triclosan bound to enoyl reductase.
J.Mol.Biol., 294, 1999
|
|
1GGN
 
 | | Structures of glycogen phosphorylase-inhibitor complexes and the implications for structure-based drug design | | Descriptor: | BETA-D-GLUCOPYRANOSE SPIROHYDANTOIN, PROTEIN (GLYCOGEN PHOSPHORYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-08-29 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranosylidene spirothiohydantoin binding to glycogen phosphorylase B.
Bioorg.Med.Chem., 10, 2002
|
|
1CPI
 
 | |
1DAN
 
 | |
