5I05
 
 | |
8CZO
 
 | | Cryo-EM structure of BCL10 CARD - MALT1 DD filament | | Descriptor: | B-cell lymphoma/leukemia 10, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | David, L, Wu, H. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | BCL10 Mutations Define Distinct Dependencies Guiding Precision Therapy for DLBCL.
Cancer Discov, 12, 2022
|
|
2BGI
 
 | | X-Ray Structure of the Ferredoxin-NADP(H) Reductase from Rhodobacter capsulatus complexed with three molecules of the detergent n-heptyl- beta-D-thioglucoside at 1.7 Angstroms | | Descriptor: | CARBON DIOXIDE, FERREDOXIN-NADP(H) REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Perez-Dorado, J.I, Hermoso, J.A, Nogues, I, Frago, S, Bittel, C, Mayhew, S.G, Gomez-Moreno, C, Medina, M, Cortez, N, Carrillo, N. | | Deposit date: | 2004-12-23 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Ferredoxin-Nadp(H) Reductase from Rhodobacter Capsulatus: Molecular Structure and Catalytic Mechanism
Biochemistry, 44, 2005
|
|
8CZD
 
 | | Cryo-EM structure of BCL10 R58Q filament | | Descriptor: | B-cell lymphoma/leukemia 10 | | Authors: | David, L, Wu, H. | | Deposit date: | 2022-05-24 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | BCL10 Mutations Define Distinct Dependencies Guiding Precision Therapy for DLBCL.
Cancer Discov, 12, 2022
|
|
5V5C
 
 | | VQIINK, Structure of the amyloid-spine from microtubule associated protein tau Repeat 2 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Seidler, P.M, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S, Cascio, D, Boyer, D.R. | | Deposit date: | 2017-03-14 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.25 Å) | | Cite: | Structure-based inhibitors of tau aggregation.
Nat Chem, 10, 2018
|
|
8D5K
 
 | |
8D5E
 
 | | The complex of Gtf2b Peptide TGAASFDEF Presented by H2-Dd | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Custodio, J.M.F, Baker, B.M. | | Deposit date: | 2022-06-04 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and physical features that distinguish tumor-controlling from inactive cancer neoepitopes.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D5F
 
 | |
8CB1
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-PNT-DNM 15 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[5-(phenanthren-9-ylmethoxy)pentyl]piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Overkleeft, H, Artola, M. | | Deposit date: | 2023-01-25 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fluorescence polarisation activity-based protein profiling for the identification of deoxynojirimycin-type inhibitors selective for lysosomal retaining alpha- and beta-glucosidases.
Chem Sci, 14, 2023
|
|
8CB6
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in covalent complex with TAMRA tagged 1,6-Epi-cylcophellitol aziridine activity based probe | | Descriptor: | (1S,2R,3R,4R,5R)-5-[8-[4-(4-azanylbutyl)-1,2,3-triazol-1-yl]octylamino]-4-(hydroxymethyl)cyclohexane-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Overkleeft, H, Artola, M. | | Deposit date: | 2023-01-25 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fluorescence polarisation activity-based protein profiling for the identification of deoxynojirimycin-type inhibitors selective for lysosomal retaining alpha- and beta-glucosidases.
Chem Sci, 14, 2023
|
|
4ZFL
 
 | |
5AYI
 
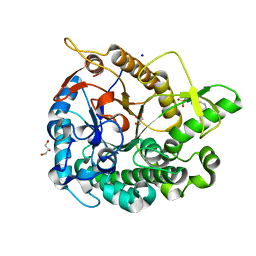 | | Crystal structure of GH1 Beta-glucosidase TD2F2 N223Q mutant | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, BETA-GLUCOSIDASE, GLYCEROL, ... | | Authors: | Jo, T, Manninen, J.A, Matsuzawa, T, Uchiyama, T, Yaoi, K, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-08-21 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and identification of a key amino acid for glucose tolerance, substrate specificity, and transglycosylation activity of metagenomic beta-glucosidase Td2F2
Febs J., 283, 2016
|
|
5UYN
 
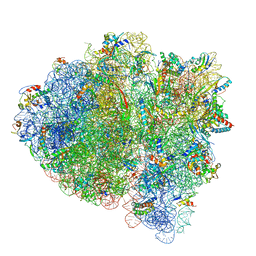 | | 70S ribosome bound with near-cognate ternary complex not base-paired to A site codon (Structure I-nc) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
5V5B
 
 | | KVQIINKKLD, Structure of the amyloid spine from microtubule associated protein tau Repeat 2 | | Descriptor: | Microtubule-associated protein tau | | Authors: | Seidler, P.M, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S, Cascio, D, Boyer, D.R. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.5 Å) | | Cite: | Structure-based inhibitors of tau aggregation.
Nat Chem, 10, 2018
|
|
5AYB
 
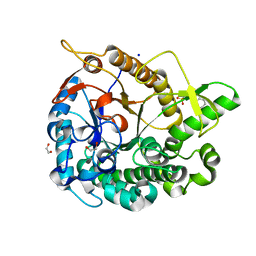 | | Crystal structure of GH1 Beta-Glucosidase TD2F2 N223G mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, BETA-GLUCOSIDASE, ... | | Authors: | Jo, T, Manninen, J.A, Matsuzawa, T, Uchiyama, T, Yaoi, K, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-08-12 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and identification of a key amino acid for glucose tolerance, substrate specificity, and transglycosylation activity of metagenomic beta-glucosidase Td2F2
Febs J., 283, 2016
|
|
8D8O
 
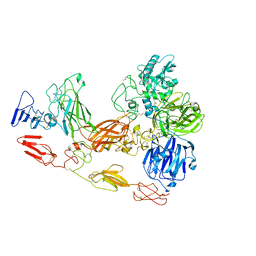 | | Cryo-EM structure of substrate unbound PAPP-A | | Descriptor: | Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
5BU6
 
 | | Structure of BpsB deaceylase domain from Bordetella bronchiseptica | | Descriptor: | 1,2-ETHANEDIOL, BpsB (PgaB), Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase, ... | | Authors: | Little, D.J, Bamford, N.C, Howell, P.L. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | The Protein BpsB Is a Poly-beta-1,6-N-acetyl-d-glucosamine Deacetylase Required for Biofilm Formation in Bordetella bronchiseptica.
J.Biol.Chem., 290, 2015
|
|
6DR7
 
 | | YopH PTP1B WPD loop Chimera 2 PTPase bound to vanadate | | Descriptor: | ACETATE ION, Targeted effector protein, VANADATE ION | | Authors: | Moise, G, Morales, Y, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | A YopH PTP1B Chimera Shows the Importance of the WPD-Loop Sequence to the Activity, Structure, and Dynamics of Protein Tyrosine Phosphatases.
Biochemistry, 57, 2018
|
|
5TD8
 
 | | Crystal structure of an Extended Dwarf Ndc80 Complex | | Descriptor: | Kinetochore protein NDC80, Kinetochore protein NUF2, Kinetochore protein SPC24, ... | | Authors: | Valverde, R, Ingram, J, Harrison, S.C. | | Deposit date: | 2016-09-17 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (7.531 Å) | | Cite: | Conserved Tetramer Junction in the Kinetochore Ndc80 Complex.
Cell Rep, 17, 2016
|
|
5W58
 
 | | Crystal Complex of Cyclooxygenase-2: (S)-ARN-2508 (a dual COX and FAAH inhibitor) | | Descriptor: | (2S)-2-{2-fluoro-3'-[(hexylcarbamoyl)oxy][1,1'-biphenyl]-4-yl}propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Goodman, M.C, Banerjee, S, Piomelli, D, Marnett, L.J. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | Dual cyclooxygenase-fatty acid amide hydrolase inhibitor exploits novel binding interactions in the cyclooxygenase active site.
J. Biol. Chem., 293, 2018
|
|
3VSB
 
 | | SUBTILISIN CARLSBERG D-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
4ZGZ
 
 | |
5A72
 
 | | Crystal structure of the homing endonuclease I-CvuI in complex with its target (Sro1.3) in the presence of 2 mM Ca | | Descriptor: | 24MER DNA, 5'-D(*DTP*CP*AP*GP*AP*AP*CP*GP*TP*CP*GP*TP*AP *DCP*GP*AP*CP*GP*TP*TP*CP*TP*GP*A)-3', CALCIUM ION, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
8D5L
 
 | | Crystal structure of theophylline aptamer in complex with TAL1 | | Descriptor: | 4-amino-8-methylpteridine-2,7(1H,8H)-dione, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Menichelli, E, Spraggon, G. | | Deposit date: | 2022-06-05 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of small molecules that target a tertiary-structured RNA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D29
 
 | |
