5ACQ
 
 | | W228A-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | BETA-LACTAMASE, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
5A87
 
 | | Crystal structure of the metallo-beta-lactamase VIM-5 | | Descriptor: | GLYCEROL, METALLO-BETA-LACTAMASE VIM-5, ZINC ION | | Authors: | Brem, J, Duzgun, A.O, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparison of Verona Integron-Borne Metallo-beta-Lactamase (VIM) Variants Reveals Differences in Stability and Inhibition Profiles.
Antimicrob. Agents Chemother., 60, 2015
|
|
5ACT
 
 | | W228S-Investigation of the impact from residues W228 and Y233 in the metallo-beta-lactamase GIM-1 | | Descriptor: | GIM-1 PROTEIN, ZINC ION | | Authors: | Skagseth, S, Carlsen, T.J, Bjerga, G.E.K, Spencer, J, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2015-08-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Role of Residues W228 and Y233 in the Structure and Activity of Metallo-Beta-Lactamase Gim-1.
Antimicrob.Agents Chemother., 60, 2015
|
|
8R5U
 
 | |
8R5T
 
 | |
8SK2
 
 | |
4NQ6
 
 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 complexed with compound L-CS319 | | Descriptor: | (3S,5S,7aR)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Beta-lactamase 2, POTASSIUM ION, ... | | Authors: | Gonzalez, J.M, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2013-11-23 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4LE6
 
 | | Crystal structure of the phosphotriesterase OPHC2 from Pseudomonas pseudoalcaligenes | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Organophosphorus hydrolase, ... | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-06-25 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Enzymatic Characterization of the Phosphotriesterase OPHC2 from Pseudomonas pseudoalcaligenes.
Plos One, 8, 2013
|
|
8SKP
 
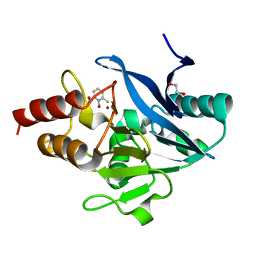 | | X-ray structure of the NDM-4 beta-lactamase from Klebsiella pneumonia in complex with 1-hydroxypyridine-2(1H)-thione-6-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 1-hydroxy-6-sulfanylidene-1,6-dihydropyridine-2-carboxylic acid, Metallo-beta-lactamase type 2, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of a novel inhibitor for the New Delhi metallo-beta-lactamase-4: Implications for drug design and combating bacterial drug resistance.
J.Biol.Chem., 299, 2023
|
|
8SKO
 
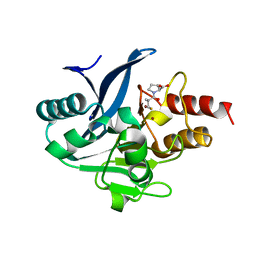 | |
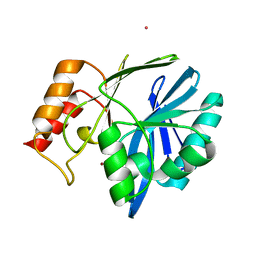
4NQ4
 
 | |
8U00
 
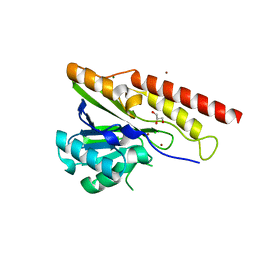 | | Crystal structure of metallo-beta-lactamase superfamily protein from Caulobacter vibrioides | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-08-28 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of metallo-beta-lactamase superfamily protein from Caulobacter vibrioides
To Be Published
|
|
4O98
 
 | | Crystal structure of Pseudomonas oleovorans PoOPH mutant H250I/I263W | | Descriptor: | ZINC ION, organophosphorus hydrolase | | Authors: | Luo, X.J, Kong, X.D, Zhao, J, Chen, Q, Zhou, J.H, Xu, J.H. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-03 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Switching a newly discovered lactonase into an efficient and thermostable phosphotriesterase by simple double mutations His250Ile/Ile263Trp
Biotechnol.Bioeng., 111, 2014
|
|
4P62
 
 | |
4NQ5
 
 | | Bacillus cereus Zn-dependent metallo-beta-lactamase at pH 7 complexed with compound CS319 | | Descriptor: | (3R,5R,7aS)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, Beta-lactamase 2, POTASSIUM ION, ... | | Authors: | Gonzalez, J.M, Gonzalez, M.M, Vila, A.J. | | Deposit date: | 2013-11-23 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Inhibition of metallo-lactamases with bicyclic compounds
TO BE PUBLISHED
|
|
4NQ2
 
 | | Structure of Zn(II)-bound metallo-beta-lactamse VIM-2 from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Aitha, M, Nix, J.C, Crowder, M.W, Page, R.C. | | Deposit date: | 2013-11-23 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical, Mechanistic, and Spectroscopic Characterization of Metallo-beta-lactamase VIM-2.
Biochemistry, 53, 2014
|
|
2WRS
 
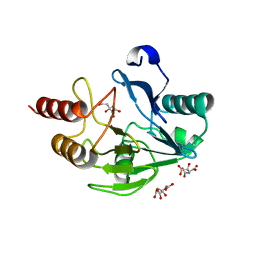 | | Crystal Structure of the Mono-Zinc Metallo-beta-lactamase VIM-4 from Pseudomonas aeruginosa | | Descriptor: | BETA-LACTAMASE VIM-4, CITRATE ANION, CITRIC ACID, ... | | Authors: | Lassaux, P, Hamel, M, Gulea, M, Delbruck, H, Traore, D.A.K, Mercuri, P.S, Horsfall, L, Dehareng, D, Gaumont, A.-C, Frere, J.-M, Ferrer, J.-L, Hoffmann, K, Galleni, M, Bebrone, C. | | Deposit date: | 2009-09-02 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Mercaptophosphonate Compounds as Broad-Spectrum Inhibitors of the Metallo-Beta-Lactamases.
J.Med.Chem., 53, 2010
|
|
4XUK
 
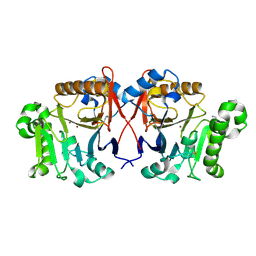 | |
3ESH
 
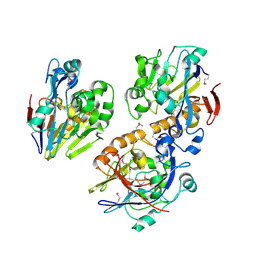 | | Crystal structure of a probable metal-dependent hydrolase from Staphylococcus aureus. Northeast Structural Genomics target ZR314 | | Descriptor: | Protein similar to metal-dependent hydrolase | | Authors: | Seetharaman, J, Chen, Y, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-06 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a probable metal-dependent hydrolase from
Staphylococcus aureus. Northeast Structural Genomics target ZR314
To be Published
|
|
2UYX
 
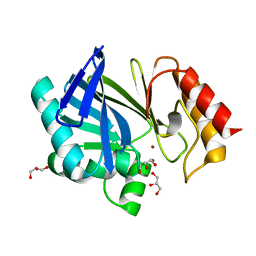 | | metallo-beta-lactamase (1BC2) single point mutant D120S | | Descriptor: | BETA-LACTAMASE II, GLYCEROL, ZINC ION | | Authors: | Larrull, L.I, Fabiane, S.M, Kowalski, J.M, Bennett, B, Sutton, B.J, Vila, A.J. | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Asp-120 locates Zn2 for optimal metallo-beta-lactamase activity.
J. Biol. Chem., 282, 2007
|
|
2VW8
 
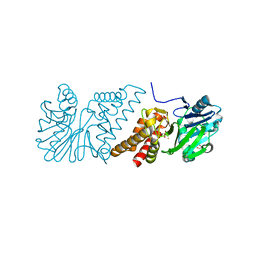 | | Crystal Structure of Quinolone signal response protein pqsE from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, FE (II) ION, ... | | Authors: | Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, Oke, M, Naismith, J.H, White, M.F. | | Deposit date: | 2008-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
3F9O
 
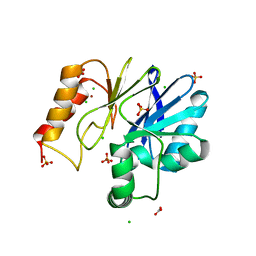 | |
1KO3
 
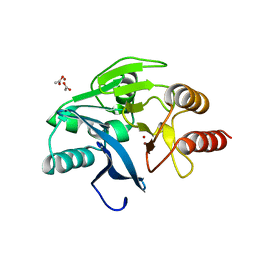 | | VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa with Cys221 reduced | | Descriptor: | ACETATE ION, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Garcia-Saez, I, Docquier, J.-D, Rossolini, G.M, Dideberg, O. | | Deposit date: | 2001-12-20 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form
J.Mol.Biol., 375, 2008
|
|
3FAI
 
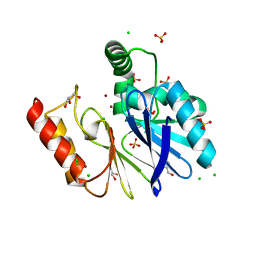 | | The Di Zinc Carbapenemase CphA N220G mutant | | Descriptor: | Beta-lactamase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Delbruck, H, Hoffmann, K.M.V. | | Deposit date: | 2008-11-17 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of the dizinc subclass B2 metallo-beta-lactamase CphA reveals that the second inhibitory zinc ion binds in the histidine site
Antimicrob.Agents Chemother., 53, 2009
|
|
3DH8
 
 | | Structure of Pseudomonas Quinolone Signal Response Protein PqsE | | Descriptor: | FE (III) ION, Uncharacterized protein PA1000, bis(4-nitrophenyl) hydrogen phosphate | | Authors: | Yu, S, Blankenfeldt, W. | | Deposit date: | 2008-06-17 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein.
Biochemistry, 48, 2009
|
|
