4GQI
 
 | | Synthesis of novel MT3 receptor ligands via unusual Knoevenagel condensation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, N-{(3R)-3-[2-(acetylamino)ethyl]-2-oxo-2,3-dihydro-1H-indol-5-yl}acetamide, ... | | Authors: | Volkova, M.S, Jensen, K.C, Lozinskaya, N.A, Sosonyuk, S.E, Proskurnina, M.V, Mesecar, A.D, Zefirov, N.S. | | Deposit date: | 2012-08-23 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Synthesis of novel МТ3 receptor ligands via an unusual Knoevenagel condensation.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2ANS
 
 | |
4GX5
 
 | | GsuK Channel | | Descriptor: | CALCIUM ION, POTASSIUM ION, TrkA domain protein, ... | | Authors: | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | Deposit date: | 2012-09-03 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
3D34
 
 | | Structure of the F-spondin domain of mindin | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Spondin-2 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-05-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the F-spondin domain of mindin, an integrin ligand and pattern recognition molecule.
Embo J., 28, 2009
|
|
2HG1
 
 | |
3NYT
 
 | | X-ray crystal structure of the WlbE (WpbE) aminotransferase from pseudomonas aeruginosa, mutation K185A, in complex with the PLP external aldimine adduct with UDP-3-amino-2-N-acetyl-glucuronic acid, at 1.3 angstrom resolution | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-6-{[(R)-{[(S)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}tetrahydro-2H-pyran-2-carboxylic acid (non-preferred name), Aminotransferase WbpE, SODIUM ION | | Authors: | Holden, H.M, Thoden, J.B. | | Deposit date: | 2010-07-15 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
1LOJ
 
 | | Crystal structure of a Methanobacterial Sm-like archaeal protein (SmAP1) bound to uridine-5'-monophosphate (UMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, URIDINE, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Mura, C, Kozhukhovsky, A, Eisenberg, D. | | Deposit date: | 2002-05-06 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs)
Protein Sci., 12, 2003
|
|
6EZX
 
 | | CATHEPSIN L IN COMPLEX WITH (3S,14E)-19-chloro-N-(1-cyanocyclopropyl)-5-oxo-17-oxa-4-azatricyclo[16.2.2.06,11]docosa-1(21),6,8,10,14,18(22),19-heptaene-3-carboxamide | | Descriptor: | (3~{S},14~{E})-19-chloranyl-~{N}-[1-(iminomethyl)cyclopropyl]-5-oxidanylidene-17-oxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6,8,10,14,18(22),19-heptaene-3-carboxamide, Cathepsin L1 | | Authors: | Banner, D.W, Benz, J, Kuglstatter, A. | | Deposit date: | 2017-11-16 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6EX8
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-9-(trifluoromethyl)-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,18(22),19-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-07 | | Release date: | 2018-04-11 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6EXQ
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-8-methoxy-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(20),6(11),7,9,18,21-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
4Z0I
 
 | | Crystal structure of a tetramer of GluA2 ligand binding domains bound with glutamate at 1.45 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, Glutamate receptor 2,Glutamate receptor 2, ... | | Authors: | Baranovic, J, Chebli, M, Salazar, H, Carbone, A.L, Ghisi, V, Faelber, K, Lau, A.Y, Daumke, O, Plested, A.J.R. | | Deposit date: | 2015-03-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the tetrameric wt GluA2 ligand-binding domain bound to glutamate at 1.45 Angstroms resolution
To Be Published
|
|
6F06
 
 | | CATHEPSIN L IN COMPLEX WITH (3S,14E)-8-(azetidin-3-yl)-19-chloro-N-(1-cyanocyclopropyl)-5-oxo-12,17-dioxa-4-azatricyclo[16.2.2.06,11]docosa-1(21),6,8,10,14,18(22),19-heptaene-3-carboxamide | | Descriptor: | (3~{S},14~{E})-8-(azetidin-3-yl)-19-chloranyl-~{N}-(1-cyanocyclopropyl)-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(21),6(11),7,9,14,18(22),19-heptaene-3-carboxamide, CHLORIDE ION, Cathepsin L1, ... | | Authors: | Kuglstatter, A, Stihle, M. | | Deposit date: | 2017-11-17 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
5UCR
 
 | |
6QZH
 
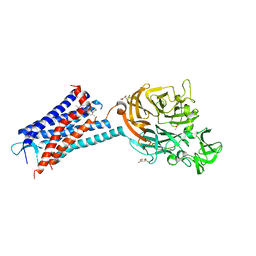 | | Structure of the human CC Chemokine Receptor 7 in complex with the intracellular allosteric antagonist Cmp2105 and the insertion protein Sialidase NanA | | Descriptor: | 3-[[4-[[(1~{R})-2,2-dimethyl-1-(5-methylfuran-2-yl)propyl]amino]-1,1-bis(oxidanylidene)-1,2,5-thiadiazol-3-yl]amino]-~{N},~{N},6-trimethyl-2-oxidanyl-benzamide, C-C chemokine receptor type 7,Sialidase A,C-C chemokine receptor type 7, D(-)-TARTARIC ACID, ... | | Authors: | Jaeger, K, Bruenle, S, Weinert, T, Guba, W, Muehle, J, Miyazaki, T, Weber, M, Furrer, A, Haenggi, N, Tetaz, T, Huang, C.Y, Mattle, D, Vonach, J.M, Gast, A, Kuglstatter, A, Rudolph, M.G, Nogly, P, Benz, J, Dawson, R.J.P, Standfuss, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Allosteric Ligand Recognition in the Human CC Chemokine Receptor 7.
Cell, 178, 2019
|
|
5H57
 
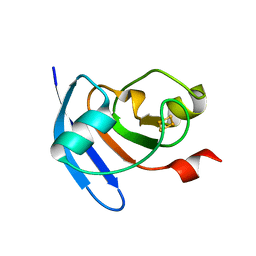 | | Ferredoxin III from maize root | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-3, chloroplastic | | Authors: | Kurisu, G, Hase, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the isotype-specific interactions of ferredoxin and ferredoxin: NADP(+) oxidoreductase: an evolutionary switch between photosynthetic and heterotrophic assimilation
Photosyn. Res., 134, 2017
|
|
1UY0
 
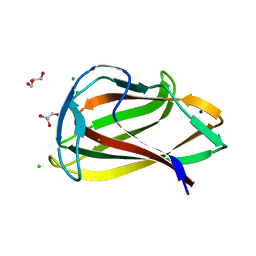 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with glc-1,3-glc-1,4-glc-1,3-glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
2KMJ
 
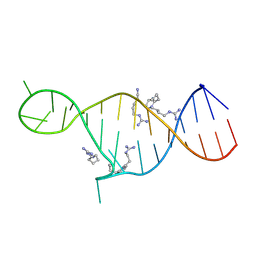 | | High resolution NMR solution structure of a complex of HIV-2 TAR RNA and a synthetic tripeptide in a 1:2 stoichiometry | | Descriptor: | Pyrimidinylpeptide, RNA (28-MER) | | Authors: | Ferner, J, Suhartono, M, Breitung, S, Jonker, H.R.A, Hennig, M, Woehnert, J, Goebel, M, Schwalbe, H. | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries.
Chembiochem, 10, 2009
|
|
1UYX
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellobiose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
7LA4
 
 | | Integrin AlphaIIbBeta3-PT25-2 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bush, M.W, Walz, T, Coller, B, Filizola, M, Spasic, A, Nesic, D, Li, J. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-12 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Electron microscopy shows that binding of monoclonal antibody PT25-2 primes integrin alpha IIb beta 3 for ligand binding.
Blood Adv, 5, 2021
|
|
1UYZ
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with xylotetraose | | Descriptor: | CALCIUM ION, CELLULASE B, GLYCEROL, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
4APF
 
 | | Crystal structure of the human KLHL11-Cul3 complex at 3.1A resolution | | Descriptor: | CULLIN 3, KELCH-LIKE PROTEIN 11 | | Authors: | Canning, P, Cooper, C.D.O, Krojer, T, Vollmar, M, Ugochukwu, E, Muniz, J.R.C, Ayinampudi, V, Savitsky, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-02 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
4XCN
 
 | | Crystal structure of human 4E10 Fab in complex with phosphatidic acid (06:0 PA); 2.9 A resolution | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 4E10 Fab heavy chain, 4E10 Fab light chain, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
9FWI
 
 | | Ensemble model of ligand-free SARS-CoV-2 NSP10-NSP14 (ExoN) and in complex with partially bound VT00025 | | Descriptor: | (3-oxidanylazetidin-1-yl)-phenyl-methanone, DIMETHYL SULFOXIDE, Guanine-N7 methyltransferase nsp14, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.533 Å) | | Cite: | Ensemble model of ligand-free SARS-CoV-2 NSP10-NSP14 (ExoN) and in complex with partially bound VT00025
To Be Published
|
|
6UC3
 
 | | Spectroscopic and structural characterization of a genetically encoded direct sensor for protein-ligand interactions | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Mills, J.H, Gleason, P.R, Simmons, C.R, Henderson, J.N, Kartchner, B.K. | | Deposit date: | 2019-09-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Origins of Altered Spectroscopic Properties upon Ligand Binding in Proteins Containing a Fluorescent Noncanonical Amino Acid.
Biochemistry, 60, 2021
|
|
4N34
 
 | | Structure of langerin CRD I313 with alpha-MeGlcNAc | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, methyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside | | Authors: | Feinberg, H, Rowntree, T.J.W, Tan, S.L.W, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Common polymorphisms in human langerin change specificity for glycan ligands.
J.Biol.Chem., 288, 2013
|
|
