1CZ7
 
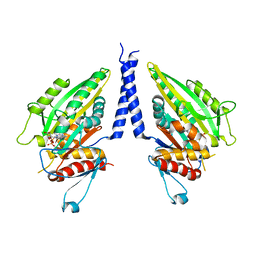 | | THE CRYSTAL STRUCTURE OF A MINUS-END DIRECTED MICROTUBULE MOTOR PROTEIN NCD REVEALS VARIABLE DIMER CONFORMATIONS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MICROTUBULE MOTOR PROTEIN NCD | | Authors: | Kozielski, F.K, De Bonis, S, Burmeister, W, Cohen-Addad, C, Wade, R. | | Deposit date: | 1999-09-01 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the minus-end-directed microtubule motor protein ncd reveals variable dimer conformations.
Structure Fold.Des., 7, 1999
|
|
3KIN
 
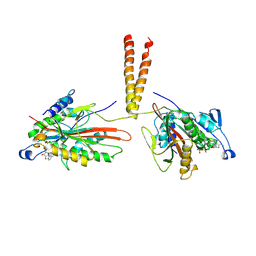 | | KINESIN (DIMERIC) FROM RATTUS NORVEGICUS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN HEAVY CHAIN | | Authors: | Kozielski, F, Sack, S, Marx, A, Thormahlen, M, Schonbrunn, E, Biou, V, Thompson, A, Mandelkow, E.-M, Mandelkow, E. | | Deposit date: | 1997-08-25 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of dimeric kinesin and implications for microtubule-dependent motility.
Cell(Cambridge,Mass.), 91, 1997
|
|
8CRK
 
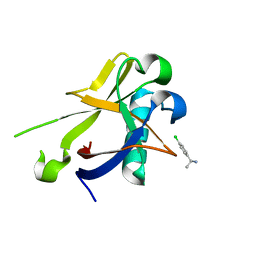 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2 refined against anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-chlorophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8CRM
 
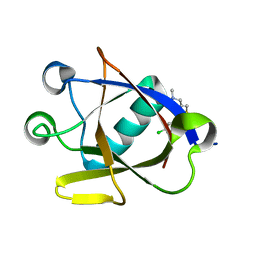 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11C6 refined against anomalous diffraction data | | Descriptor: | 1-[2-(3-chlorophenyl)-1,3-thiazol-4-yl]-~{N}-methyl-methanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8CRF
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 5E11 refined against anomalous diffraction data | | Descriptor: | Host translation inhibitor nsp1, ~{N}-methyl-1-(4-thiophen-2-ylphenyl)methanamine | | Authors: | Ma, S, Mykhaylyk, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8BZN
 
 | | SARS-CoV-2 non-structural protein 10 (nsp10) variant T102I | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab, ... | | Authors: | Wang, H, Rizvi, S.R.A, Dong, D, Lou, J, Wang, Q, Sopipong, W, Najar, F, Agarwal, P.K, Kozielski, F, Haider, S. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Emerging variants of SARS-CoV-2 NSP10 highlight strong functional conservation of its binding to two non-structural proteins, NSP14 and NSP16.
Elife, 12, 2023
|
|
8A4Y
 
 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with N-(2,3-dihydro-1H-inden-5-yl)acetamide | | Descriptor: | Host translation inhibitor nsp1, N-(2,3-dihydro-1H-inden-5-yl)acetamide | | Authors: | Borsatto, A, Galdadas, I, Ma, S, Damfo, S, Haider, S, Kozielski, F, Estarellas, C, Gervasio, F.L. | | Deposit date: | 2022-06-13 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Revealing druggable cryptic pockets in the Nsp1 of SARS-CoV-2 and other beta-coronaviruses by simulations and crystallography.
Elife, 11, 2022
|
|
8AYS
 
 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 4-(2-aminothiazol-4-yl)phenol | | Descriptor: | 4-(2-amino-1,3-thiazol-4-yl)phenol, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
2E9S
 
 | | human neuronal Rab6B in three intermediate forms | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Vellieux, F.M, Tcherniuk, S, Garcia-Saez, I, Kozielski, F. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 3D structure of human neuronal Rab6B in three intermediate forms
To be Published
|
|
5NJV
 
 | | Flavivirus NS5 domain | | Descriptor: | CHLORIDE ION, NS5, S-ADENOSYLMETHIONINE | | Authors: | Talapatra, S.K, Chatrin, C, Kozielski, F. | | Deposit date: | 2017-03-29 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of the binary methyltransferase-SAH complex from Zika virus reveals a novel conformation for the mechanism of mRNA capping.
Oncotarget, 9, 2018
|
|
5NJU
 
 | | Flavivirus NS5 domain | | Descriptor: | Genome polyprotein, MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Talapatra, S.K, Chatrin, C, Kozielski, F. | | Deposit date: | 2017-03-29 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the binary methyltransferase-SAH complex from Zika virus reveals a novel conformation for the mechanism of mRNA capping.
Oncotarget, 9, 2018
|
|
8A55
 
 | |
8AZ8
 
 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 2-(benzylamino)ethan-1-ol | | Descriptor: | 2-[(phenylmethyl)amino]ethanol, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2022-09-05 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
6H9R
 
 | | Dengue-RdRp3-inhibitor complex soaking | | Descriptor: | 2-(4-methoxy-3-thiophen-2-yl-phenyl)ethanoic acid, DI(HYDROXYETHYL)ETHER, Genome polyprotein, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development and validation of RdRp Screen, a crystallization screen for viral RNA-dependent RNA polymerases.
Biol Open, 8, 2019
|
|
6HKY
 
 | | Eg5-inhibitor complex | | Descriptor: | (2~{S})-2-(3-azanylpropyl)-5-[2,5-bis(fluoranyl)phenyl]-~{N}-methoxy-~{N}-methyl-2-phenyl-1,3,4-thiadiazole-3-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-09-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Is the Fate of Clinical Candidate Arry-520 Already Sealed? Predicting Resistance in Eg5-Inhibitor Complexes.
Mol.Cancer Ther., 18, 2019
|
|
6HKX
 
 | | Eg5-inhibitor complex | | Descriptor: | (2~{S})-2-(3-azanylpropyl)-5-[2,5-bis(fluoranyl)phenyl]-~{N}-methoxy-~{N}-methyl-2-phenyl-1,3,4-thiadiazole-3-carboxamide, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-09-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Is the Fate of Clinical Candidate Arry-520 Already Sealed? Predicting Resistance in Eg5-Inhibitor Complexes.
Mol.Cancer Ther., 18, 2019
|
|
1T5C
 
 | | Crystal structure of the motor domain of human kinetochore protein CENP-E | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Centromeric protein E, MAGNESIUM ION, ... | | Authors: | Garcia-Saez, I, Yen, T, Wade, R.H, Kozielski, F, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-05-04 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the motor domain of the human kinetochore protein CENP-E.
J.Mol.Biol., 340, 2004
|
|
6Y1U
 
 | | Mycobacterium tuberculosis FtsZ-GDP in complex with 4-hydroxycoumarin | | Descriptor: | 4-HYDROXY-2H-CHROMEN-2-ONE, Cell division protein FtsZ, DIMETHYL SULFOXIDE, ... | | Authors: | Alnami, A.T, Norton, R.S, Pena, H.P, Haider, M, kozielski, F. | | Deposit date: | 2020-02-13 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conformational Flexibility of A Highly Conserved Helix Controls Cryptic Pocket Formation in FtsZ.
J.Mol.Biol., 433, 2021
|
|
6Y1V
 
 | | Mycobacterium tuberculosis FtsZ-GTP-gamma-S in complex with 4-hydroxycoumarin | | Descriptor: | 4-HYDROXY-2H-CHROMEN-2-ONE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein FtsZ, ... | | Authors: | Alnami, A.T, Norton, R.S, Pena, H.P, Haider, M, kozielski, F. | | Deposit date: | 2020-02-13 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Flexibility of A Highly Conserved Helix Controls Cryptic Pocket Formation in FtsZ.
J.Mol.Biol., 433, 2021
|
|
6ZPE
 
 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Replicase polyprotein 1ab, ... | | Authors: | Fisher, S.Z, Kozielski, F. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
2IEH
 
 | |
6G6Z
 
 | | Eg5-inhibitor complex | | Descriptor: | (~{N}~{Z})-~{N}-[(5~{S})-4-ethanoyl-5-methyl-5-phenyl-1,3,4-thiadiazolidin-2-ylidene]ethanamide, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Talapatra, S.K, Kozielski, F, Tham, C.L. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Eg5 - K858 complex and implications for structure-based design of thiadiazole-containing inhibitors.
Eur J Med Chem, 156, 2018
|
|
6YM1
 
 | | Mycobacterium tuberculosis FtsZ in complex with GDP | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Alnami, A.T, Norton, R.S, Pena, H.P, Haider, M, kozielski, F. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational Flexibility of A Highly Conserved Helix Controls Cryptic Pocket Formation in FtsZ.
J.Mol.Biol., 433, 2021
|
|
6YM9
 
 | | Mycobacterium tuberculosis FtsZ in complex with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein FtsZ | | Authors: | Alnami, A.T, Norton, R.S, Pena, H.P, Haider, M, kozielski, F. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Conformational Flexibility of A Highly Conserved Helix Controls Cryptic Pocket Formation in FtsZ.
J.Mol.Biol., 433, 2021
|
|
2KIN
 
 | | KINESIN (MONOMERIC) FROM RATTUS NORVEGICUS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN, SULFATE ION | | Authors: | Sack, S, Muller, J, Kozielski, F, Marx, A, Thormahlen, M, Biou, V, Mandelkow, E.-M, Brady, S.T, Mandelkow, E. | | Deposit date: | 1997-04-28 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of motor and neck domains from rat brain kinesin.
Biochemistry, 36, 1997
|
|
