3NNZ
 
 | | Structure of rat neuronal nitric oxide synthase heme domain complexed with 6-(((3S,4S)-4-(2-(3-Fluorophenethylamino)ethoxy)pyrrolidin-3-yl)methyl)pyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3S,4S)-4-(2-{[2-(3-fluorophenyl)ethyl]amino}ethoxy)pyrrolidin-3-yl]methyl}pyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2010-06-24 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Peripheral but crucial: a hydrophobic pocket (Tyr(706), Leu(337), and Met(336)) for potent and selective inhibition of neuronal nitric oxide synthase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NPZ
 
 | | Prolactin Receptor (PRLR) Complexed with the Natural Hormone (PRL) | | Descriptor: | Prolactin, Prolactin receptor | | Authors: | Van Agthoven, J, England, P, Goffin, V, Broutin, I. | | Deposit date: | 2010-06-29 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural characterization of the stem-stem dimerization interface between prolactin receptor chains complexed with the natural hormone.
J.Mol.Biol., 404, 2010
|
|
6ZBV
 
 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor and sybody Sb_GlyT1#7 | | Descriptor: | Sodium- and chloride-dependent glycine transporter 1,Sodium- and chloride-dependent glycine transporter 1, Sybody Sb_GlyT1#7, [5-fluoranyl-6-(oxan-4-yloxy)-1,3-dihydroisoindol-2-yl]-[5-methylsulfonyl-2-[2,2,3,3,3-pentakis(fluoranyl)propoxy]phenyl]methanone | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-06-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
1IMI
 
 | | SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1 | | Descriptor: | PROTEIN (ALPHA-CONOTOXIN IMI) | | Authors: | Maslennikov, I.V, Shenkarev, Z.O, Zhmak, M.N, Tsetlin, V.I, Ivanov, V.T, Arseniev, A.S. | | Deposit date: | 1998-11-27 | | Release date: | 1999-04-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR spatial structure of alpha-conotoxin ImI reveals a common scaffold in snail and snake toxins recognizing neuronal nicotinic acetylcholine receptors.
FEBS Lett., 444, 1999
|
|
3NSE
 
 | | BOVINE ENOS, H4B-FREE, SEITU COMPLEX | | Descriptor: | ACETATE ION, ARGININE, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1998-09-23 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center.
Cell(Cambridge,Mass.), 95, 1998
|
|
3OH6
 
 | | AlkA Undamaged DNA Complex: Interrogation of a C:G base pair | | Descriptor: | 5'-D(*GP*AP*CP*AP*(BRU)P*GP*AP*AP*(BRU)P*GP*CP*C)-3', 5'-D(*GP*CP*AP*TP*TP*CP*AP*TP*GP*TP*C)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
J.Biol.Chem., 285, 2010
|
|
8IED
 
 | | Cryo-EM structure of GPR156-miniGo-scFv16 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Shin, J, Park, J, Cho, Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Constitutive activation mechanism of a class C GPCR.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6ZIE
 
 | | Crystal structure of MCL-1 in complex with a neutralizing Alphabody CMPX-383B | | Descriptor: | CMPX-383B, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Pannecoucke, E, Savvides, S.N, Desmet, J, Lasters, I. | | Deposit date: | 2020-06-25 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cell-penetrating Alphabody protein scaffolds for intracellular drug targeting.
Sci Adv, 7, 2021
|
|
8IY9
 
 | | Structure of Niacin-GPR109A-G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8IU2
 
 | | Cryo-EM structure of Long-wave-sensitive opsin 1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Peng, Q, Cheng, X.Y, Li, J, Lu, Q.Y, Li, Y.Y, Zhang, J. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of Long-wave-sensitive opsin 1
To Be Published
|
|
3ODK
 
 | | Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-pyridin-2-yl-1H-pyrazole-5-carboxylic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8IYH
 
 | | Structure of MK6892-GPR109A-G-protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8IYW
 
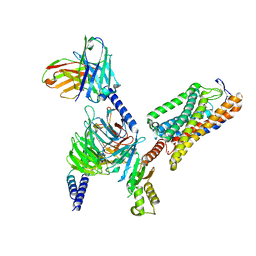 | | Structure of GSK256073-GPR109A-G-protein complex | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
1DK5
 
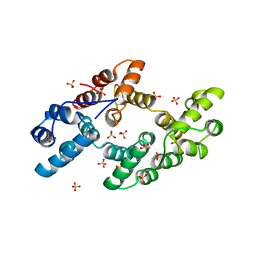 | | CRYSTAL STRUCTURE OF ANNEXIN 24(CA32) FROM CAPSICUM ANNUUM | | Descriptor: | ANNEXIN 24(CA32), SULFATE ION | | Authors: | Hofmann, A, Proust, J, Dorowski, A, Schantz, R, Huber, R. | | Deposit date: | 1999-12-06 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Annexin 24 from Capsicum annuum. X-ray structure and biochemical characterization.
J.Biol.Chem., 275, 2000
|
|
8IZ4
 
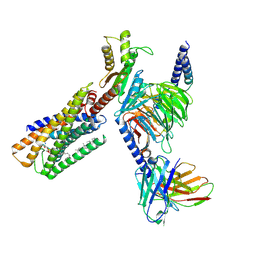 | | Lysophosphatidylserine receptor GPR34-Gi complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Gong, W, Liu, G, Li, X, Zhang, X. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural mechanisms of ligand binding and signaling in lysophosphatidylserine receptors
To Be Published
|
|
3O0R
 
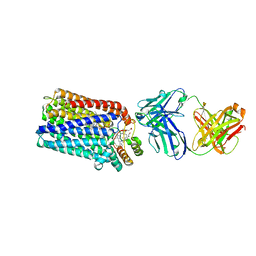 | | Crystal structure of nitric oxide reductase from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, FE (III) ION, HEME C, ... | | Authors: | Hino, T, Matsumoto, Y, Nagano, S, Sugimoto, H, Fukumori, Y, Murata, T, Iwata, S, Shiro, Y. | | Deposit date: | 2010-07-20 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of biological N2O generation by bacterial nitric oxide reductase
Science, 330, 2010
|
|
8J6I
 
 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound MK-6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-04-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
8J6L
 
 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound niacin | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-04-03 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
1GV0
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
8J23
 
 | | Cryo-EM structure of FFAR2 complex in apo state | | Descriptor: | Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short chain fatty acid receptors FFAR2 and FFAR3
To Be Published
|
|
3OJX
 
 | | Disulfide crosslinked cytochrome P450 reductase inactive | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Hamdane, D, Shen, A, Choi, V, Kasper, C, Zhang, H, Im, S.-C, Waskell, L, Kim, J.-J.P. | | Deposit date: | 2010-08-23 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Changes of NADPH-Cytochrome P450 Oxidoreductase Are Essential for Catalysis and Cofactor Binding.
J.Biol.Chem., 286, 2011
|
|
3OMA
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1GXR
 
 | | WD40 Region of Human Groucho/TLE1 | | Descriptor: | CALCIUM ION, TRANSDUCIN-LIKE ENHANCER PROTEIN 1 | | Authors: | Pearl, L.H, Roe, S.M, Pickles, L.M. | | Deposit date: | 2002-04-10 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the C-Terminal Wd40 Repeat Domain of the Human Groucho/Tle1 Transcriptional Corepressor
Structure, 10, 2002
|
|
1HF4
 
 | | STRUCTURAL EFFECTS OF MONOVALENT ANIONS ON POLYMORPHIC LYSOZYME CRYSTALS | | Descriptor: | LYSOZYME, NITRATE ION, SODIUM ION | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 2000-11-29 | | Release date: | 2001-01-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Effects of Monovalent Anions on Polymorphic Lysozyme Crystals
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GXW
 
 | | the 2.2 A resolution structure of thermolysin crystallized in presence of potassium thiocyanate | | Descriptor: | CALCIUM ION, LYSINE, THERMOLYSIN, ... | | Authors: | Gaucher, J.F, Selkti, M, Prange, T, Tomas, A. | | Deposit date: | 2002-04-12 | | Release date: | 2002-12-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The 2.2 A Resolution Structure of Thermolysin (Tln) Crystallized in the Presence of Potassium Thiocyanate.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
