7K19
 
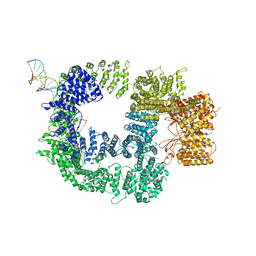 | | CryoEM structure of DNA-PK catalytic subunit complexed with DNA (Complex I) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K1J
 
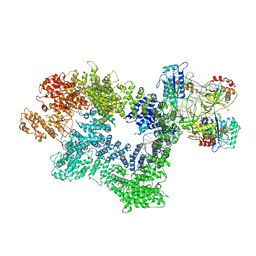 | | CryoEM structure of inactivated-form DNA-PK (Complex III) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
5WVK
 
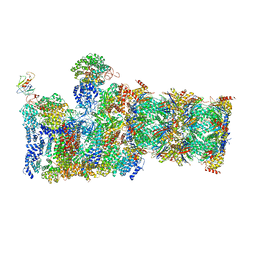 | | Yeast proteasome-ADP-AlFx | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
5D3G
 
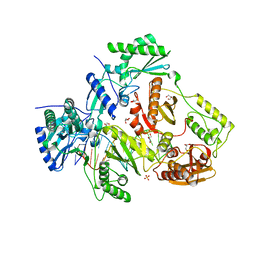 | | Structure of HIV-1 Reverse Transcriptase Bound to a Novel 38-mer Hairpin Template-Primer DNA Aptamer | | Descriptor: | DNA aptamer (38-MER), GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer.
Protein Sci., 25, 2016
|
|
5WVI
 
 | | The resting state of yeast proteasome | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Ding, Z, Cong, Y. | | Deposit date: | 2016-12-25 | | Release date: | 2017-03-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | High-resolution cryo-EM structure of the proteasome in complex with ADP-AlFx
Cell Res., 27, 2017
|
|
7K1N
 
 | | CryoEM structure of inactivated-form DNA-PK (Complex V) | | Descriptor: | DNA (5'-D(P*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-08 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
5ZBB
 
 | | Crystal structure of Rtt109-Asf1-H3-H4 complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
7OWG
 
 | | human DEPTOR in a complex with mutant human mTORC1 A1459P | | Descriptor: | DEP domain-containing mTOR-interacting protein, Regulatory-associated protein of mTOR, Serine/threonine-protein kinase mTOR, ... | | Authors: | Heimhalt, M, Berndt, A, Wagstaff, J, Anandapadamanaban, M, Perisic, O, Maslen, S, McLaughlin, S, Yu, W.-H, Masson, G.R, Boland, A, Ni, X, Yamashita, K, Murshudov, G.N, Skehel, M, Freund, S.M, Williams, R.L. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Bipartite binding and partial inhibition links DEPTOR and mTOR in a mutually antagonistic embrace.
Elife, 10, 2021
|
|
7K1K
 
 | | CryoEM structure of inactivated-form DNA-PK (Complex IV) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
3ZRC
 
 | | pVHL54-213-EloB-EloC complex (4R)-4-HYDROXY-1-[(3-METHYLISOXAZOL-5-YL)ACETYL]-N-[4-(1,3-OXAZOL-5-YL)BENZYL]-L-PROLINAMIDE bound | | Descriptor: | (4R)-4-HYDROXY-1-[(3-METHYLISOXAZOL-5-YL)ACETYL]-N-[4-(1,3-OXAZOL-5-YL)BENZYL]-L-PROLINAMIDE, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 2, ... | | Authors: | Van Molle, I, Buckley, D.L, Crews, C.M, Ciulli, A. | | Deposit date: | 2011-06-15 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Targeting the Von Hippel-Lindau E3 Ubiquitin Ligase Using Small Molecules to Disrupt the Vhl/Hif-1Alpha Interaction
J.Am.Chem.Soc., 134, 2012
|
|
5HRO
 
 | |
3ZRF
 
 | | pVHL54-213-EloB-EloC complex_apo | | Descriptor: | TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 2, VON HIPPEL-LINDAU DISEASE TUMOR SUPPRESSOR, | | Authors: | Van Molle, I, Buckley, D.L, Crews, C.M, Ciulli, A. | | Deposit date: | 2011-06-16 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeting the Von Hippel-Lindau E3 Ubiquitin Ligase Using Small Molecules to Disrupt the Vhl/Hif-1Alpha Interaction
J.Am.Chem.Soc., 134, 2012
|
|
6O9O
 
 | | Crystal Structure of SMYD3 with Potent and Selective Isoxazole Amide Inhibitor 1 | | Descriptor: | 5-cyclopropyl-N-{1-[({trans-4-[(4,4,4-trifluorobutyl)amino]cyclohexyl}methyl)sulfonyl]piperidin-4-yl}-1,2-oxazole-3-carboxamide, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Elkins, P.A, Wang, L. | | Deposit date: | 2019-03-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Discovery of Isoxazole Amides as Potent and Selective SMYD3 Inhibitors
To be published
|
|
3HVT
 
 | | STRUCTURAL BASIS OF ASYMMETRY IN THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE HETERODIMER | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Steitz, T.A, Smerdon, S.J, Jaeger, J, Wang, J, Kohlstaedt, L.A, Chirino, A.J, Friedman, J.M, Rice, P.A. | | Deposit date: | 1994-07-25 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.Usa, 91, 1994
|
|
8H2H
 
 | | Cryo-EM structure of a Group II Intron Complexed with its Reverse Transcriptase | | Descriptor: | Group II intron-encoded protein LtrA, LtrB, RNA (5'-R(P*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*C)-3') | | Authors: | Liu, N, Dong, X.L, Qu, G.S, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2022-10-06 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Functionalized graphene grids with various charges for single-particle cryo-EM.
Nat Commun, 13, 2022
|
|
6P6G
 
 | | Co-crystal Structure of human SMYD3 with Isoxazole Amides Inhibitors | | Descriptor: | 5-cyclopropyl-N-{1-[({trans-4-[(4,4,4-trifluorobutyl)amino]cyclohexyl}methyl)sulfonyl]piperidin-4-yl}-1,2-oxazole-3-carboxamide, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Elkins, P.A, Wang, L. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Isoxazole Amides as Potent and Selective SMYD3 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6P6K
 
 | |
7NTF
 
 | | Cryo-EM structure of unliganded O-GlcNAc transferase | | Descriptor: | Isoform 1 of UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Meek, R.W, Blaza, J.N, Davies, G.J. | | Deposit date: | 2021-03-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.32 Å) | | Cite: | Cryo-EM structure provides insights into the dimer arrangement of the O-linked beta-N-acetylglucosamine transferase OGT.
Nat Commun, 12, 2021
|
|
7UX2
 
 | |
6P7Z
 
 | |
4WZS
 
 | |
6PAF
 
 | |
3BAP
 
 | |
5LQF
 
 | | CDK1/CyclinB1/CKS2 in complex with NU6102 | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B.J, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E, Newell, D.R, Turner, D.M, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Endicott, J.A, Cano, C. | | Deposit date: | 2016-08-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
6ZFP
 
 | | Cryo-EM structure of DNA-PKcs (State 2) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
