6V9I
 
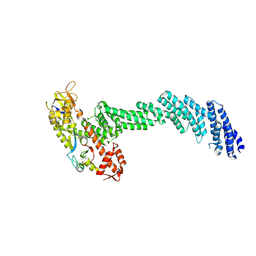 | | cryo-EM structure of Cullin5 bound to RING-box protein 2 (Cul5-Rbx2) | | Descriptor: | Immunoglobulin G-binding protein G,Cullin-5, RING-box protein 2, ZINC ION | | Authors: | Komives, E.A, Lumpkin, R.J, Baker, R.W, Leschziner, A.E. | | Deposit date: | 2019-12-13 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structure and dynamics of the ASB9 CUL-RING E3 Ligase.
Nat Commun, 11, 2020
|
|
8CIL
 
 | |
3V7D
 
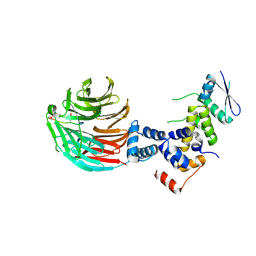 | | Crystal Structure of ScSkp1-ScCdc4-pSic1 peptide complex | | Descriptor: | Cell division control protein 4, Protein SIC1, Suppressor of kinetochore protein 1 | | Authors: | Tang, X, Orlicky, S, Mittag, T, Csizmok, V, Pawson, T, Forman-Kay, J, Sicheri, F, Tyers, M. | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3GCB
 
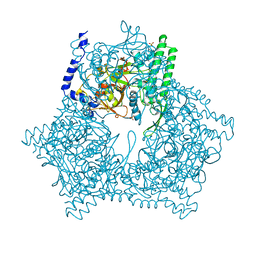 | | GAL6 (YEAST BLEOMYCIN HYDROLASE) MUTANT C73A/DELTAK454 | | Descriptor: | GAL6, GLYCEROL, SULFATE ION | | Authors: | Joshua-Tor, L, Zheng, W, Johnston, S.A. | | Deposit date: | 1998-02-27 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The unusual active site of Gal6/bleomycin hydrolase can act as a carboxypeptidase, aminopeptidase, and peptide ligase.
Cell(Cambridge,Mass.), 93, 1998
|
|
6SLM
 
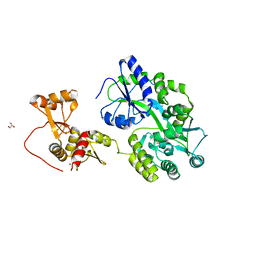 | | Crystal structure of full-length HPV31 E6 oncoprotein in complex with LXXLL peptide of ubiquitin ligase E6AP | | Descriptor: | GLYCEROL, Maltose/maltodextrin-binding periplasmic protein,Protein E6,Ubiquitin-protein ligase E3A, ZINC ION, ... | | Authors: | Conrady, M, Gogl, G, Cousido-Siah, A, Mitschler, A, Trave, G, Simon, C. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of High-Risk Papillomavirus 31 E6 Oncogenic Protein and Characterization of E6/E6AP/p53 Complex Formation.
J.Virol., 95, 2020
|
|
8WQD
 
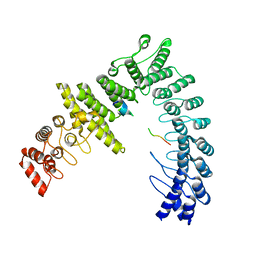 | |
8WQI
 
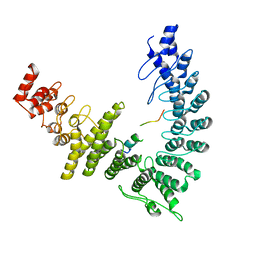 | |
8JE2
 
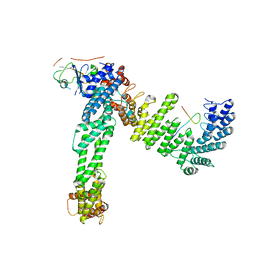 | | Cryo-EM structure of neddylated Cul2-Rbx1-EloBC-FEM1B complexed with FNIP1-FLCN | | Descriptor: | Cullin-2, Elongin-B, Elongin-C, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
4GIZ
 
 | | Crystal structure of full-length human papillomavirus oncoprotein E6 in complex with LXXLL peptide of ubiquitin ligase E6AP at 2.55 A resolution | | Descriptor: | Maltose-binding periplasmic protein, UBIQUITIN LIGASE EA6P: chimeric protein, Protein E6, ... | | Authors: | McEwen, A.G, Zanier, K, Charbonnier, S, Poussin, P, Cura, V, Vande Pol, S, Trave, G, Cavarelli, J. | | Deposit date: | 2012-08-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins.
Science, 339, 2013
|
|
2BAY
 
 | | Crystal structure of the Prp19 U-box dimer | | Descriptor: | Pre-mRNA splicing factor PRP19 | | Authors: | Vander Kooi, C.W, Ohi, M.D, Rosenberg, J.A, Oldham, M.L, Newcomer, M.E, Gould, K.L, Chazin, W.J. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases.
Biochemistry, 45, 2006
|
|
7BY1
 
 | |
2Y43
 
 | |
3MKS
 
 | | Crystal Structure of yeast Cdc4/Skp1 in complex with an allosteric inhibitor SCF-I2 | | Descriptor: | 1,1'-binaphthalene-2,2'-dicarboxylic acid, Cell division control protein 4, GLYCEROL, ... | | Authors: | Orlicky, S, Sicheri, F, Tyers, M, Tang, X. | | Deposit date: | 2010-04-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase.
Nat.Biotechnol., 28, 2010
|
|
8OKX
 
 | | Structure of cGAS in complex with SPSB3-ELOBC | | Descriptor: | Cyclic GMP-AMP synthase, Elongin-B, Elongin-C, ... | | Authors: | Xu, P.B, Ablasser, A. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | The CRL5-SPSB3 ubiquitin ligase targets nuclear cGAS for degradation.
Nature, 627, 2024
|
|
4FJO
 
 | | Structure of the Rev1 CTD-Rev3/7-Pol kappa RIR complex | | Descriptor: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | Wojtaszek, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa
J.Biol.Chem., 287, 2012
|
|
2A4D
 
 | | Structure of the human ubiquitin-conjugating enzyme E2 variant 1 (UEV-1) | | Descriptor: | Ubiquitin-conjugating enzyme E2 variant 1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-28 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
4H2X
 
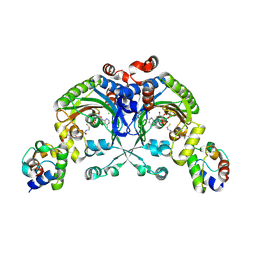 | | Crystal structure of engineered Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with carrier protein from Agrobacterium tumefaciens and an analogue of glycyl adenylate | | Descriptor: | 4'-PHOSPHOPANTETHEINE, 5'-O-(glycylsulfamoyl)adenosine, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
6VE5
 
 | |
6OAA
 
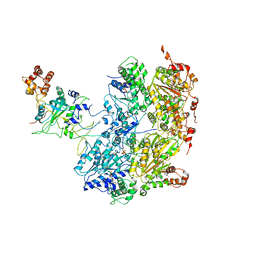 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ADP-BeFx, state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6OA9
 
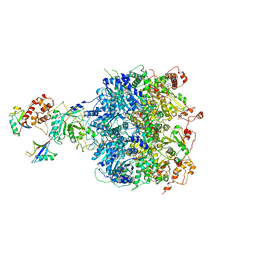 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
6LTV
 
 | | Crystal Structure of I122A/I330A variant of S-adenosylmethionine synthetase from Cryptosporidium hominis in complex with ONB-SAM (2-nitro benzyme S-adenosyl-methionine) | | Descriptor: | MAGNESIUM ION, S-adenosylmethionine synthase, TRIPHOSPHATE, ... | | Authors: | Singh, R.K, Michailidou, F, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Engineered SAM Synthetases for Enzymatic Generation of AdoMet Analogs with Photocaging Groups and Reversible DNA Modification in Cascade Reactions.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6LTW
 
 | | Crystal structure of Apo form of I122A/I330A variant of S-adenosylmethionine synthetase from Cryptosporidium hominis | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, S-adenosylmethionine synthase | | Authors: | Singh, R.K, Michailidou, F, Rentmeister, A, Kuemmel, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineered SAM Synthetases for Enzymatic Generation of AdoMet Analogs with Photocaging Groups and Reversible DNA Modification in Cascade Reactions.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8A58
 
 | |
7P3A
 
 | | N-terminal domain of CGI-99 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Kroupova, A, Jinek, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular architecture of the human tRNA ligase complex.
Elife, 10, 2021
|
|
7JL0
 
 | | Cryo-EM structure of MDA5-dsRNA in complex with TRIM65 PSpry domain (Monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
