8C33
 
 | |
8C37
 
 | |
8C31
 
 | |
6D27
 
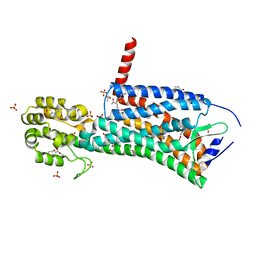 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with CAY10471 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, OLEIC ACID, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
6SOF
 
 | | human insulin receptor ectodomain bound by 4 insulin | | Descriptor: | Insulin, Insulin receptor | | Authors: | Gutmann, T, Schaefer, I.B, Poojari, C.S, Vattulainen, I, Strauss, M, Coskun, U. | | Deposit date: | 2019-08-29 | | Release date: | 2019-11-13 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain.
J.Cell Biol., 219, 2020
|
|
3M7R
 
 | | Crystal structure of VDR H305Q mutant | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Vitamin D3 receptor | | Authors: | Rochel, N, Hourai, S, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hereditary vitamin D-resistant rickets--associated mutant H305Q of vitamin D nuclear receptor bound to its natural ligand
J.Steroid Biochem.Mol.Biol., 121, 2010
|
|
1NHZ
 
 | | Crystal Structure of the Antagonist Form of Glucocorticoid Receptor | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, GLUCOCORTICOID RECEPTOR, HEXANE-1,6-DIOL | | Authors: | Kauppi, B, Jakob, C, Farnegardh, M, Yang, J, Ahola, H, Alarcon, M, Calles, K, Engstrom, O, Harlan, J, Muchmore, S, Ramqvist, A.-K, Thorell, S, Ohman, L, Greer, J, Gustafsson, J.-A, Carlstedt-Duke, J, Carlquist, M. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Three-dimensional Structures of Antagonistic and Agonistic Forms of the Glucocorticoid Receptor Ligand-binding Domain:
RU-486 INDUCES A TRANSCONFORMATION THAT LEADS TO ACTIVE ANTAGONISM.
J.Biol.Chem., 278, 2003
|
|
5JL7
 
 | |
8QMI
 
 | | Crystal structure of RNA G2C4 repeats - native model | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | Authors: | Blaszczyk, L, Kiliszek, A. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 52, 2024
|
|
1NBP
 
 | | Crystal Structure Of Human Interleukin-2 Y31C Covalently Modified At C31 With 3-Mercapto-1-(1,3,4,9-tetrahydro-B-carbolin-2-yl)-propan-1-one | | Descriptor: | 3-MERCAPTO-1-(1,3,4,9-TETRAHYDRO-B-CARBOLIN-2-YL)-PROPAN-1-ONE, Interleukin-2, SULFATE ION | | Authors: | Hyde, J, Braisted, A.C, Randal, M, Arkin, M.R. | | Deposit date: | 2002-12-03 | | Release date: | 2002-12-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and characterization of cooperative ligand binding in the adaptive region of interleukin-2
Biochemistry, 42, 2003
|
|
8C3B
 
 | | X-ray structure of RNase A upon reaction with a Ruthenium(II)-arene Complexed with Glycosylated Carbene Ligands (5) | | Descriptor: | (1,3-dimethyl-2~{H}-imidazol-2-yl)-oxidanyl-oxidanylidene-ruthenium, (1,3-dimethylimidazol-1-ium-2-yl)-tetrakis(oxidanyl)ruthenium, (1,3-dimethylimidazol-1-ium-2-yl)-tris(oxidanyl)ruthenium, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Ruthenium(II)–Arene Complexes with Glycosylated NHC-Carbene Co-Ligands: Synthesis, Hydrolytic Behavior, and Binding to Biological Molecules
Organometallics, 42, 2023
|
|
8C39
 
 | | X-ray structure of HEWL upon reaction with a Ruthenium(II)-arene Complexed with Glycosylated Carbene Ligands (5) | | Descriptor: | CHLORIDE ION, Lysozyme, NITRATE ION, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ruthenium(II)–Arene Complexes with Glycosylated NHC-Carbene Co-Ligands: Synthesis, Hydrolytic Behavior, and Binding to Biological Molecules
Organometallics, 42, 2023
|
|
6C3Y
 
 | | Wild type structure of SiRHP | | Descriptor: | IRON/SULFUR CLUSTER, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Stroupe, M.E. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.542 Å) | | Cite: | The role of extended Fe4S4cluster ligands in mediating sulfite reductase hemoprotein activity.
Biochim. Biophys. Acta, 1866, 2018
|
|
5JL6
 
 | |
5JL9
 
 | |
2RKM
 
 | | STRUCTURE OF OPPA COMPLEXED WITH LYS-LYS | | Descriptor: | LYSINE, OLIGO-PEPTIDE BINDING PROTEIN, URANYL (VI) ION | | Authors: | Sleigh, S.H, Tame, J.R.H, Wilkinson, A.J. | | Deposit date: | 1997-03-25 | | Release date: | 1997-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine.
Biochemistry, 36, 1997
|
|
5N5T
 
 | | 14-3-3 sigma in complex with TAZ pS89 peptide and fragment NV2 | | Descriptor: | 14-3-3 protein sigma, 4-[3,5-bis(chloranyl)pyridin-2-yl]oxyphenol, CHLORIDE ION, ... | | Authors: | Sijbesma, E, Leysen, S, Ottmann, C. | | Deposit date: | 2017-02-14 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening.
Biochemistry, 56, 2017
|
|
3H0F
 
 | | Crystal structure of the human Fyn SH3 R96W mutant | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Proto-oncogene tyrosine-protein kinase Fyn | | Authors: | Ponchon, L, Hoh, F, Labesse, G, Dumas, C, Arold, S.T. | | Deposit date: | 2009-04-09 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Synergy and allostery in ligand binding by HIV-1 Nef.
Biochem.J., 478, 2021
|
|
3H0I
 
 | |
3VJS
 
 | | Vitamin D receptor complex with a carborane compound | | Descriptor: | 1-(2-[(S)-2,4-Dihydroxybutoxy]ethyl)-12-(5-ethyl-5-hydroxyheptyl)-1,12-dicarba-closo-dodecaborane, Vitamin D3 receptor, peptide from Mediator of RNA polymerase II transcription subunit 1 | | Authors: | Fujii, S, Masuno, M, Kagechika, H, Nakabayashi, M, Ito, N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Boron Cluster-based Development of Potent Nonsecosteroidal Vitamin D Receptor Ligands: Direct Observation of Hydrophobic Interaction between Protein Surface and Carborane
J.Am.Chem.Soc., 133, 2011
|
|
3MB9
 
 | | Human Aldose Reductase mutant T113A complexed with Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2010-03-25 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ligand-induced fit affects binding modes and provokes changes in crystal packing of aldose reductase
Biochim.Biophys.Acta, 1810, 2011
|
|
8R1I
 
 | |
7ENV
 
 | | crystal structure of NS5 in complex with the N-terminal bromodomain of BRD2 (BRD2-BD1). | | Descriptor: | 7-chloranyl-2-[(3-chlorophenyl)amino]pyrano[3,4-e][1,3]oxazine-4,5-dione, Bromodomain-containing protein 2, SULFATE ION | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S. | | Deposit date: | 2021-04-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural investigation of a pyrano-1,3-oxazine derivative and the phenanthridinone core moiety against BRD2 bromodomains.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7EO5
 
 | | Crystal structure of pyrano 1,3, oxazine derivative in complex with the second bromodomain of BRD2 | | Descriptor: | 7-chloranyl-2-[(3-chlorophenyl)amino]pyrano[3,4-e][1,3]oxazine-4,5-dione, Bromodomain-containing protein 2, TRIETHYLENE GLYCOL | | Authors: | Padmanabhan, B, Arole, A, Deshmukh, P, Ashok, S. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of a pyrano-1,3-oxazine derivative and the phenanthridinone core moiety against BRD2 bromodomains.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
3C5E
 
 | | Crystal structure of human acyl-CoA synthetase medium-chain family member 2A (L64P mutation) in complex with ATP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A synthetase ACSM2A, ... | | Authors: | Pilka, E.S, Kochan, G.T, Bhatia, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl-coenzyme A synthetase ACSM2A.
J.Mol.Biol., 388, 2009
|
|
