4J1W
 
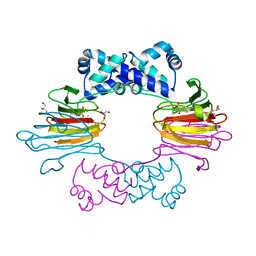 | |
4O75
 
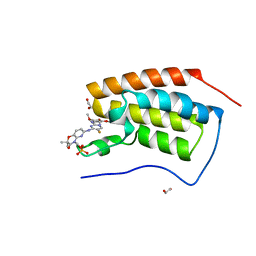 | | Crystal structure of the first bromodomain of human BRD4 in complex with FOSTAMATINIB | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, [6-({5-fluoro-2-[(3,4,5-trimethoxyphenyl)amino]pyrimidin-4-yl}amino)-2,2-dimethyl-3-oxo-2,3-dihydro-4H-pyrido[3,2-b][1,4]oxazin-4-yl]methyl dihydrogen phosphate | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
5JQ6
 
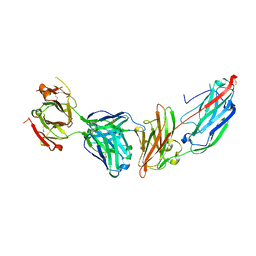 | | Crystal structure of ClfA in complex with the Fab fragment of Tefibazumab | | Descriptor: | Clumping factor A, Tefibazumab FAB FRAGMENT HEAVY CHAIN, Tefibazumab FAB FRAGMENT LIGHT CHAIN | | Authors: | Ganesh, V.K. | | Deposit date: | 2016-05-04 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lessons from the Crystal Structure of the S. aureus Surface Protein Clumping Factor A in Complex With Tefibazumab, an Inhibiting Monoclonal Antibody.
EBioMedicine, 13, 2016
|
|
7S5A
 
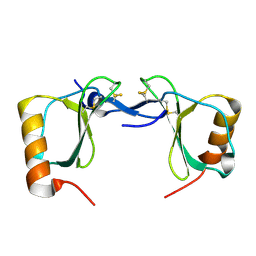 | | Crystal structure of human chemokine CCL8 | | Descriptor: | C-C motif chemokine 8 | | Authors: | Bhusal, R.P, Devkota, S.R, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2021-09-10 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure-guided engineering of tick evasins for targeting chemokines in inflammatory diseases.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5KBX
 
 | |
8RDU
 
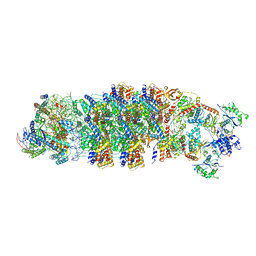 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K composite map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LE, MAGNESIUM ION, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
8RKU
 
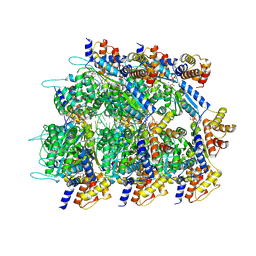 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsC domain local-refinement map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-target strand - LE, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
8RKV
 
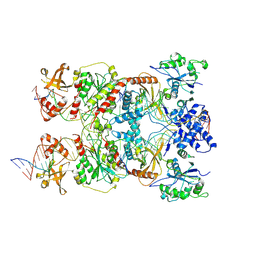 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsB domain local-refinement map | | Descriptor: | LE, MAGNESIUM ION, Non-target strand - LE, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
8RKT
 
 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K Cas12k domain local-refinement map | | Descriptor: | MAGNESIUM ION, Non-target strand - LE, ShCas12k, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
3UPY
 
 | | Crystal structure of the Brucella abortus enzyme catalyzing the first committed step of the methylerythritol 4-phosphate pathway. | | Descriptor: | 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MAGNESIUM ION, Oxidoreductase | | Authors: | Calisto, B.M, Perez-Gil, J, Fita, I, Rodriguez-Concepcion, M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Brucella abortus deoxyxylulose-5-phosphate reductoisomerase-like (DRL) enzyme involved in isoprenoid biosynthesis.
J.Biol.Chem., 287, 2012
|
|
7BFZ
 
 | | X-ray structure of human prostate-specific membrane antigen(PSMA) in complex with a inhibitor Glu-490 | | Descriptor: | (((S)-1-carboxy-5-((E)-2-cyano-3-(5-(1-(3-methoxy-3-oxopropyl)-1,2,3,4-tetrahydroquinolin-6-yl)thiophen-2-yl)acrylamido)pentyl)carbamoyl)-L-glutamic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rakhimbekova, A, Motlova, L, Barinka, C. | | Deposit date: | 2021-01-05 | | Release date: | 2021-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A prostate-specific membrane antigen activated molecular rotor for real-time fluorescence imaging.
Nat Commun, 12, 2021
|
|
3QS7
 
 | |
3QS9
 
 | |
4V1H
 
 | |
3EIG
 
 | |
6B41
 
 | | Menin bound to M-525 | | Descriptor: | Menin, methyl {(1S,2R)-2-[(S)-cyano[1-({1-[4-({1-[4-(dimethylamino)butanoyl]azetidin-3-yl}sulfonyl)phenyl]azetidin-3-yl}methyl)piperidin-4-yl](3-fluorophenyl)methyl]cyclopentyl}carbamate, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-09-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design of the First-in-Class, Highly Potent Irreversible Inhibitor Targeting the Menin-MLL Protein-Protein Interaction.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7Z16
 
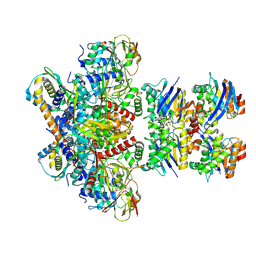 | | E. coli C-P lyase bound to PhnK/PhnL dual ABC dimer with AMPPNP and PhnK E171Q mutation | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnI, ... | | Authors: | Amstrup, S.K, Sofus, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
3SWE
 
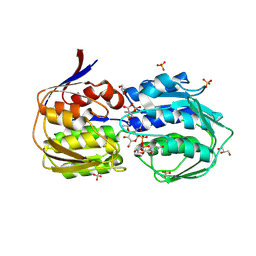 | | Haemophilus influenzae MurA in complex with UDP-N-acetylmuramic acid and covalent adduct of PEP with Cys117 | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-07-13 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
7UJD
 
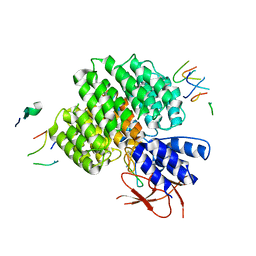 | | PSMD2 Structure bound to MC1 and Fab8/14 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 2, ACY-PHE-PRO-ASP-VAL-SAR-LEU-HIS-ARG-TYR-TRP-GLY-TRP-ASP-CYS-GLY-NH2, Fab 14 HC CDRs, ... | | Authors: | Johnson, M.C, Bashore, C, Ciferri, C, Dueber, E.C. | | Deposit date: | 2022-03-30 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Targeted degradation via direct 26S proteasome recruitment.
Nat.Chem.Biol., 19, 2023
|
|
7UIH
 
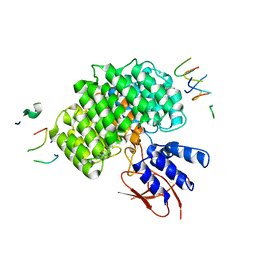 | | PSMD2 Structure | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 2, Fab 14 HC CDRs, Fab 14 LC CDRs, ... | | Authors: | Johnson, M.C, Bashore, C, Ciferri, C, Dueber, E.C. | | Deposit date: | 2022-03-29 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Targeted degradation via direct 26S proteasome recruitment.
Nat.Chem.Biol., 19, 2023
|
|
5CDM
 
 | | 2.5A structure of QPT-1 with S.aureus DNA gyrase and DNA | | Descriptor: | (2R,4S,4aS)-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4',6'(1'H,3'H)-trione, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
7UAD
 
 | | Crystal structure of human PTPN2 with inhibitor ABBV-CLS-484 | | Descriptor: | 5-{(7R)-1-fluoro-3-hydroxy-7-[(3-methylbutyl)amino]-5,6,7,8-tetrahydronaphthalen-2-yl}-1lambda~6~,2,5-thiadiazolidine-1,1,3-trione, Tyrosine-protein phosphatase non-receptor type 2 | | Authors: | Longenecker, K.L, Qiu, W, Sun, Q, Frost, J.M. | | Deposit date: | 2022-03-12 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | The PTPN2/PTPN1 inhibitor ABBV-CLS-484 unleashes potent anti-tumour immunity.
Nature, 622, 2023
|
|
5CDN
 
 | | 2.8A structure of etoposide with S.aureus DNA gyrase and DNA | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP**GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*C*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*C)-3'), ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDQ
 
 | | 2.95A structure of Moxifloxacin with S.aureus DNA gyrase and DNA | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5CDO
 
 | | 3.15A structure of QPT-1 with S.aureus DNA gyrase and DNA | | Descriptor: | (2R,4S,4aS)-4',6'-dihydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidin]-2'-one, (2R,4S,4aS,5R)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, (2R,4S,4aS,5S)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
